EMD-0689
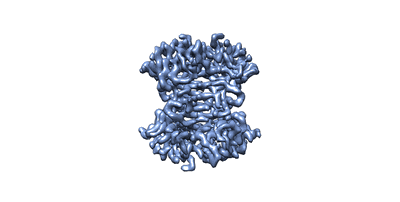
The reconstruction of biotin-bound streptavidin at 3.2 Angstrom resolution
EMD-0689
Single-particle3.2 Å
 Deposition: 15/01/2019
Deposition: 15/01/2019Map released: 29/05/2019
Last modified: 27/03/2024
Sample Organism:
Streptomyces avidinii
Sample: Streptavidin with biotin
Fitted models: 6j6j (Avg. Q-score: 0.517)
Raw data: EMPIAR-10270
Deposition Authors: Fan X ,
Wang J,
Lei JL,
Wang HW
,
Wang J,
Lei JL,
Wang HW 
Sample: Streptavidin with biotin
Fitted models: 6j6j (Avg. Q-score: 0.517)
Raw data: EMPIAR-10270
Deposition Authors: Fan X
 ,
Wang J,
Lei JL,
Wang HW
,
Wang J,
Lei JL,
Wang HW 
Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.
Fan X  ,
Wang J,
Zhang X,
Yang Z,
Zhang JC,
Zhao L,
Peng HL
,
Wang J,
Zhang X,
Yang Z,
Zhang JC,
Zhao L,
Peng HL  ,
Lei J
,
Lei J  ,
Wang HW
,
Wang HW 
(2019) Nat Commun , 10 , 2386 - 2386
 ,
Wang J,
Zhang X,
Yang Z,
Zhang JC,
Zhao L,
Peng HL
,
Wang J,
Zhang X,
Yang Z,
Zhang JC,
Zhao L,
Peng HL  ,
Lei J
,
Lei J  ,
Wang HW
,
Wang HW 
(2019) Nat Commun , 10 , 2386 - 2386
Abstract:
The fast development of single-particle cryogenic electron microscopy (cryo-EM) has made it more feasible to obtain the 3D structure of well-behaved macromolecules with a molecular weight higher than 300 kDa at ~3 Å resolution. However, it remains a challenge to obtain the high-resolution structures of molecules smaller than 200 kDa using single-particle cryo-EM. In this work, we apply the Cs-corrector-VPP-coupled cryo-EM to study the 52 kDa streptavidin (SA) protein supported on a thin layer of graphene and embedded in vitreous ice. We are able to solve both the apo-SA and biotin-bound SA structures at near-atomic resolution using single-particle cryo-EM. We demonstrate that the method has the potential to determine the structures of molecules as small as 39 kDa.
The fast development of single-particle cryogenic electron microscopy (cryo-EM) has made it more feasible to obtain the 3D structure of well-behaved macromolecules with a molecular weight higher than 300 kDa at ~3 Å resolution. However, it remains a challenge to obtain the high-resolution structures of molecules smaller than 200 kDa using single-particle cryo-EM. In this work, we apply the Cs-corrector-VPP-coupled cryo-EM to study the 52 kDa streptavidin (SA) protein supported on a thin layer of graphene and embedded in vitreous ice. We are able to solve both the apo-SA and biotin-bound SA structures at near-atomic resolution using single-particle cryo-EM. We demonstrate that the method has the potential to determine the structures of molecules as small as 39 kDa.
