EMD-11721
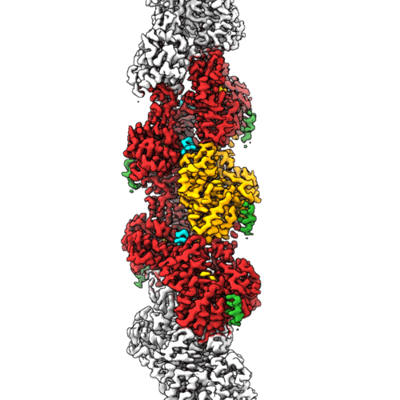
Structure of the Lifeact-F-actin complex
EMD-11721
Helical reconstruction3.5 Å
 Deposition: 14/09/2020
Deposition: 14/09/2020Map released: 28/10/2020
Last modified: 02/12/2020
Sample Organism:
Saccharomyces cerevisiae,
Rabbit,
synthetic construct
Sample: Complex between skeletal Actin from rabbit, Phalloidin and LifeAct
Fitted models: 7ad9 (Avg. Q-score: 0.466)
Deposition Authors: Belyy A, Merino F, Sitsel O, Raunser S
Sample: Complex between skeletal Actin from rabbit, Phalloidin and LifeAct
Fitted models: 7ad9 (Avg. Q-score: 0.466)
Deposition Authors: Belyy A, Merino F, Sitsel O, Raunser S
Structure of the Lifeact-F-actin complex.
Abstract:
Lifeact is a short actin-binding peptide that is used to visualize filamentous actin (F-actin) structures in live eukaryotic cells using fluorescence microscopy. However, this popular probe has been shown to alter cellular morphology by affecting the structure of the cytoskeleton. The molecular basis for such artefacts is poorly understood. Here, we determined the high-resolution structure of the Lifeact-F-actin complex using electron cryo-microscopy (cryo-EM). The structure reveals that Lifeact interacts with a hydrophobic binding pocket on F-actin and stretches over 2 adjacent actin subunits, stabilizing the DNase I-binding loop (D-loop) of actin in the closed conformation. Interestingly, the hydrophobic binding site is also used by actin-binding proteins, such as cofilin and myosin and actin-binding toxins, such as the hypervariable region of TccC3 (TccC3HVR) from Photorhabdus luminescens and ExoY from Pseudomonas aeruginosa. In vitro binding assays and activity measurements demonstrate that Lifeact indeed competes with these proteins, providing an explanation for the altering effects of Lifeact on cell morphology in vivo. Finally, we demonstrate that the affinity of Lifeact to F-actin can be increased by introducing mutations into the peptide, laying the foundation for designing improved actin probes for live cell imaging.
Lifeact is a short actin-binding peptide that is used to visualize filamentous actin (F-actin) structures in live eukaryotic cells using fluorescence microscopy. However, this popular probe has been shown to alter cellular morphology by affecting the structure of the cytoskeleton. The molecular basis for such artefacts is poorly understood. Here, we determined the high-resolution structure of the Lifeact-F-actin complex using electron cryo-microscopy (cryo-EM). The structure reveals that Lifeact interacts with a hydrophobic binding pocket on F-actin and stretches over 2 adjacent actin subunits, stabilizing the DNase I-binding loop (D-loop) of actin in the closed conformation. Interestingly, the hydrophobic binding site is also used by actin-binding proteins, such as cofilin and myosin and actin-binding toxins, such as the hypervariable region of TccC3 (TccC3HVR) from Photorhabdus luminescens and ExoY from Pseudomonas aeruginosa. In vitro binding assays and activity measurements demonstrate that Lifeact indeed competes with these proteins, providing an explanation for the altering effects of Lifeact on cell morphology in vivo. Finally, we demonstrate that the affinity of Lifeact to F-actin can be increased by introducing mutations into the peptide, laying the foundation for designing improved actin probes for live cell imaging.

