EMD-12860
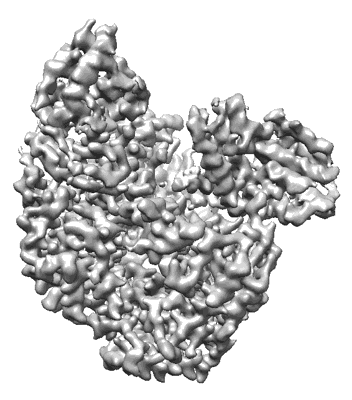
Apo-structure of Lassa virus L protein (well-resolved endonuclease) [APO-ENDO]
EMD-12860
Single-particle3.35 Å
 Deposition: 01/05/2021
Deposition: 01/05/2021Map released: 01/12/2021
Last modified: 23/10/2024
Sample Organism:
Lassa mammarenavirus
Sample: RNA-directed RNA polymerase L Lassa mammarenavirus
Fitted models: 7oe3 (Avg. Q-score: 0.456)
Deposition Authors: Kouba T ,
Vogel D
,
Vogel D 
Sample: RNA-directed RNA polymerase L Lassa mammarenavirus
Fitted models: 7oe3 (Avg. Q-score: 0.456)
Deposition Authors: Kouba T
 ,
Vogel D
,
Vogel D 
Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Kouba T  ,
Vogel D
,
Vogel D  ,
Thorkelsson SR
,
Thorkelsson SR  ,
Quemin ERJ
,
Quemin ERJ  ,
Williams HM
,
Williams HM  ,
Milewski M
,
Milewski M  ,
Busch C,
Gunther S
,
Busch C,
Gunther S  ,
Grunewald K
,
Grunewald K  ,
Rosenthal M
,
Rosenthal M  ,
Cusack S
,
Cusack S 
(2021) Nat Commun , 12 , 7018 - 7018
 ,
Vogel D
,
Vogel D  ,
Thorkelsson SR
,
Thorkelsson SR  ,
Quemin ERJ
,
Quemin ERJ  ,
Williams HM
,
Williams HM  ,
Milewski M
,
Milewski M  ,
Busch C,
Gunther S
,
Busch C,
Gunther S  ,
Grunewald K
,
Grunewald K  ,
Rosenthal M
,
Rosenthal M  ,
Cusack S
,
Cusack S 
(2021) Nat Commun , 12 , 7018 - 7018
Abstract:
Lassa virus is endemic in West Africa and can cause severe hemorrhagic fever. The viral L protein transcribes and replicates the RNA genome via its RNA-dependent RNA polymerase activity. Here, we present nine cryo-EM structures of the L protein in the apo-, promoter-bound pre-initiation and active RNA synthesis states. We characterize distinct binding pockets for the conserved 3' and 5' promoter RNAs and show how full-promoter binding induces a distinct pre-initiation conformation. In the apo- and early elongation states, the endonuclease is inhibited by two distinct L protein peptides, whereas in the pre-initiation state it is uninhibited. In the early elongation state, a template-product duplex is bound in the active site cavity together with an incoming non-hydrolysable nucleotide and the full C-terminal region of the L protein, including the putative cap-binding domain, is well-ordered. These data advance our mechanistic understanding of how this flexible and multifunctional molecular machine is activated.
Lassa virus is endemic in West Africa and can cause severe hemorrhagic fever. The viral L protein transcribes and replicates the RNA genome via its RNA-dependent RNA polymerase activity. Here, we present nine cryo-EM structures of the L protein in the apo-, promoter-bound pre-initiation and active RNA synthesis states. We characterize distinct binding pockets for the conserved 3' and 5' promoter RNAs and show how full-promoter binding induces a distinct pre-initiation conformation. In the apo- and early elongation states, the endonuclease is inhibited by two distinct L protein peptides, whereas in the pre-initiation state it is uninhibited. In the early elongation state, a template-product duplex is bound in the active site cavity together with an incoming non-hydrolysable nucleotide and the full C-terminal region of the L protein, including the putative cap-binding domain, is well-ordered. These data advance our mechanistic understanding of how this flexible and multifunctional molecular machine is activated.
