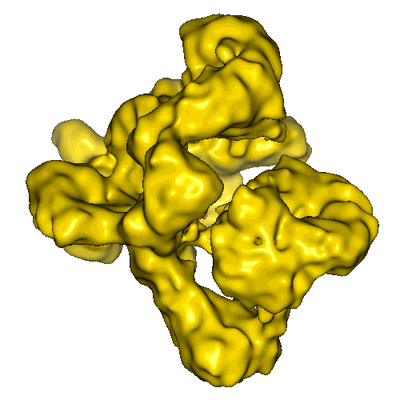
EMD-1802
Conformational flexibility of RNA polymerase III during transcriptional elongation
EMD-1802
Single-particle11.5 Å
 Deposition: 04/10/2010
Deposition: 04/10/2010Map released: 01/11/2010
Last modified: 09/09/2011
Sample Organism:
Saccharomyces cerevisiae
Sample: RNA Polymerase III
Deposition Authors: Fernandez-Tornero C ,
Bottcher B
,
Bottcher B  ,
Rashid UJ,
Steuerwald U,
Florchinger B,
Devos DP
,
Rashid UJ,
Steuerwald U,
Florchinger B,
Devos DP  ,
Lindner D,
Muller CW
,
Lindner D,
Muller CW 
Sample: RNA Polymerase III
Deposition Authors: Fernandez-Tornero C
 ,
Bottcher B
,
Bottcher B  ,
Rashid UJ,
Steuerwald U,
Florchinger B,
Devos DP
,
Rashid UJ,
Steuerwald U,
Florchinger B,
Devos DP  ,
Lindner D,
Muller CW
,
Lindner D,
Muller CW 
Conformational flexibility of RNA polymerase III during transcriptional elongation.
Fernandez-Tornero C  ,
Bottcher B
,
Bottcher B  ,
Rashid UJ,
Steuerwald U,
Florchinger B,
Devos DP
,
Rashid UJ,
Steuerwald U,
Florchinger B,
Devos DP  ,
Lindner D,
Muller CW
,
Lindner D,
Muller CW 
(2010) Embo J. , 29 , 3762 - 3772
 ,
Bottcher B
,
Bottcher B  ,
Rashid UJ,
Steuerwald U,
Florchinger B,
Devos DP
,
Rashid UJ,
Steuerwald U,
Florchinger B,
Devos DP  ,
Lindner D,
Muller CW
,
Lindner D,
Muller CW 
(2010) Embo J. , 29 , 3762 - 3772
Abstract:
RNA polymerase (Pol) III is responsible for the transcription of genes encoding small RNAs, including tRNA, 5S rRNA and U6 RNA. Here, we report the electron cryomicroscopy structures of yeast Pol III at 9.9 Å resolution and its elongation complex at 16.5 Å resolution. Particle sub-classification reveals prominent EM densities for the two Pol III-specific subcomplexes, C31/C82/C34 and C37/C53, that can be interpreted using homology models. While the winged-helix-containing C31/C82/C34 subcomplex initiates transcription from one side of the DNA-binding cleft, the C37/C53 subcomplex accesses the transcription bubble from the opposite side of this cleft. The transcribing Pol III enzyme structure not only shows the complete incoming DNA duplex, but also reveals the exit path of newly synthesized RNA. During transcriptional elongation, the Pol III-specific subcomplexes tightly enclose the incoming DNA duplex, which likely increases processivity and provides structural insights into the conformational switch between Pol III-mediated initiation and elongation.
RNA polymerase (Pol) III is responsible for the transcription of genes encoding small RNAs, including tRNA, 5S rRNA and U6 RNA. Here, we report the electron cryomicroscopy structures of yeast Pol III at 9.9 Å resolution and its elongation complex at 16.5 Å resolution. Particle sub-classification reveals prominent EM densities for the two Pol III-specific subcomplexes, C31/C82/C34 and C37/C53, that can be interpreted using homology models. While the winged-helix-containing C31/C82/C34 subcomplex initiates transcription from one side of the DNA-binding cleft, the C37/C53 subcomplex accesses the transcription bubble from the opposite side of this cleft. The transcribing Pol III enzyme structure not only shows the complete incoming DNA duplex, but also reveals the exit path of newly synthesized RNA. During transcriptional elongation, the Pol III-specific subcomplexes tightly enclose the incoming DNA duplex, which likely increases processivity and provides structural insights into the conformational switch between Pol III-mediated initiation and elongation.
