EMD-18415
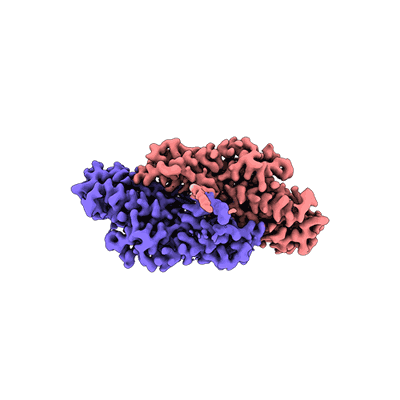
Cysteine tRNA ligase homodimer
EMD-18415
Single-particle2.8 Å
 Deposition: 08/09/2023
Deposition: 08/09/2023Map released: 29/11/2023
Last modified: 28/02/2024
Sample Organism:
Pyrococcus furiosus DSM 3638
Sample: Homodimeric form of Cysteine tRNA ligase
Fitted models: 8qhp (Avg. Q-score: 0.546)
Deposition Authors: Pacesa M ,
Correia BE
,
Correia BE  ,
Levy ED
,
Levy ED 
Sample: Homodimeric form of Cysteine tRNA ligase
Fitted models: 8qhp (Avg. Q-score: 0.546)
Deposition Authors: Pacesa M
 ,
Correia BE
,
Correia BE  ,
Levy ED
,
Levy ED 
An atlas of protein homo-oligomerization across domains of life.
Schweke H  ,
Pacesa M
,
Pacesa M  ,
Levin T,
Goverde CA
,
Levin T,
Goverde CA  ,
Kumar P
,
Kumar P  ,
Duhoo Y,
Dornfeld LJ
,
Duhoo Y,
Dornfeld LJ  ,
Dubreuil B,
Georgeon S,
Ovchinnikov S,
Woolfson DN
,
Dubreuil B,
Georgeon S,
Ovchinnikov S,
Woolfson DN  ,
Correia BE
,
Correia BE  ,
Dey S,
Levy ED
,
Dey S,
Levy ED 
(2024) Cell , 187 , 999
 ,
Pacesa M
,
Pacesa M  ,
Levin T,
Goverde CA
,
Levin T,
Goverde CA  ,
Kumar P
,
Kumar P  ,
Duhoo Y,
Dornfeld LJ
,
Duhoo Y,
Dornfeld LJ  ,
Dubreuil B,
Georgeon S,
Ovchinnikov S,
Woolfson DN
,
Dubreuil B,
Georgeon S,
Ovchinnikov S,
Woolfson DN  ,
Correia BE
,
Correia BE  ,
Dey S,
Levy ED
,
Dey S,
Levy ED 
(2024) Cell , 187 , 999
Abstract:
Protein structures are essential to understanding cellular processes in molecular detail. While advances in artificial intelligence revealed the tertiary structure of proteins at scale, their quaternary structure remains mostly unknown. We devise a scalable strategy based on AlphaFold2 to predict homo-oligomeric assemblies across four proteomes spanning the tree of life. Our results suggest that approximately 45% of an archaeal proteome and a bacterial proteome and 20% of two eukaryotic proteomes form homomers. Our predictions accurately capture protein homo-oligomerization, recapitulate megadalton complexes, and unveil hundreds of homo-oligomer types, including three confirmed experimentally by structure determination. Integrating these datasets with omics information suggests that a majority of known protein complexes are symmetric. Finally, these datasets provide a structural context for interpreting disease mutations and reveal coiled-coil regions as major enablers of quaternary structure evolution in human. Our strategy is applicable to any organism and provides a comprehensive view of homo-oligomerization in proteomes.
Protein structures are essential to understanding cellular processes in molecular detail. While advances in artificial intelligence revealed the tertiary structure of proteins at scale, their quaternary structure remains mostly unknown. We devise a scalable strategy based on AlphaFold2 to predict homo-oligomeric assemblies across four proteomes spanning the tree of life. Our results suggest that approximately 45% of an archaeal proteome and a bacterial proteome and 20% of two eukaryotic proteomes form homomers. Our predictions accurately capture protein homo-oligomerization, recapitulate megadalton complexes, and unveil hundreds of homo-oligomer types, including three confirmed experimentally by structure determination. Integrating these datasets with omics information suggests that a majority of known protein complexes are symmetric. Finally, these datasets provide a structural context for interpreting disease mutations and reveal coiled-coil regions as major enablers of quaternary structure evolution in human. Our strategy is applicable to any organism and provides a comprehensive view of homo-oligomerization in proteomes.
