EMD-23371
Polyclonal Immune complex of Fab binding to BG505 SOSIP.v4.1 from serum of rabbit 2425 at wk 18
EMD-23371
Single-particle20.0 Å
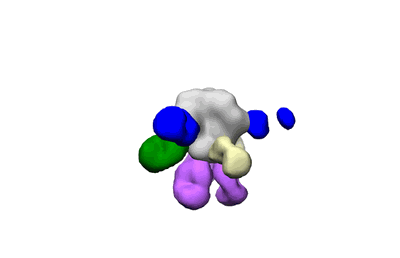 Deposition: 26/01/2021
Deposition: 26/01/2021Map released: 10/03/2021
Last modified: 10/03/2021
Sample Organism:
Oryctolagus cuniculus,
Human immunodeficiency virus
Sample: Polyclonal Immune complex of Fab binding to BG505 SOSIP.v4.1 from serum of rabbit 2425 at wk 18
Deposition Authors: Pratap PP, Ward AB
Sample: Polyclonal Immune complex of Fab binding to BG505 SOSIP.v4.1 from serum of rabbit 2425 at wk 18
Deposition Authors: Pratap PP, Ward AB
High-resolution mapping of the neutralizing and binding specificities of polyclonal sera post-HIV Env trimer vaccination.
Dingens AS  ,
Pratap P
,
Pratap P  ,
Malone K,
Hilton SK,
Ketas T,
Cottrell CA,
Overbaugh J
,
Malone K,
Hilton SK,
Ketas T,
Cottrell CA,
Overbaugh J  ,
Moore JP,
Klasse PJ
,
Moore JP,
Klasse PJ  ,
Ward AB
,
Ward AB  ,
Bloom JD
,
Bloom JD 
(2021) eLife , 10
 ,
Pratap P
,
Pratap P  ,
Malone K,
Hilton SK,
Ketas T,
Cottrell CA,
Overbaugh J
,
Malone K,
Hilton SK,
Ketas T,
Cottrell CA,
Overbaugh J  ,
Moore JP,
Klasse PJ
,
Moore JP,
Klasse PJ  ,
Ward AB
,
Ward AB  ,
Bloom JD
,
Bloom JD 
(2021) eLife , 10
Abstract:
Mapping polyclonal serum responses is critical to rational vaccine design. However, most high-resolution mapping approaches involve isolating and characterizing individual antibodies, which incompletely defines the polyclonal response. Here we use two complementary approaches to directly map the specificities of the neutralizing and binding antibodies of polyclonal anti-HIV-1 sera from rabbits immunized with BG505 Env SOSIP trimers. We used mutational antigenic profiling to determine how all mutations in Env affected viral neutralization and electron microscopy polyclonal epitope mapping (EMPEM) to directly visualize serum Fabs bound to Env trimers. The dominant neutralizing specificities were generally only a subset of the more diverse binding specificities. Additional differences between binding and neutralization reflected antigenicity differences between virus and soluble Env trimer. Furthermore, we refined residue-level epitope specificity directly from sera, revealing subtle differences across sera. Together, mutational antigenic profiling and EMPEM yield a holistic view of the binding and neutralizing specificity of polyclonal sera.
Mapping polyclonal serum responses is critical to rational vaccine design. However, most high-resolution mapping approaches involve isolating and characterizing individual antibodies, which incompletely defines the polyclonal response. Here we use two complementary approaches to directly map the specificities of the neutralizing and binding antibodies of polyclonal anti-HIV-1 sera from rabbits immunized with BG505 Env SOSIP trimers. We used mutational antigenic profiling to determine how all mutations in Env affected viral neutralization and electron microscopy polyclonal epitope mapping (EMPEM) to directly visualize serum Fabs bound to Env trimers. The dominant neutralizing specificities were generally only a subset of the more diverse binding specificities. Additional differences between binding and neutralization reflected antigenicity differences between virus and soluble Env trimer. Furthermore, we refined residue-level epitope specificity directly from sera, revealing subtle differences across sera. Together, mutational antigenic profiling and EMPEM yield a holistic view of the binding and neutralizing specificity of polyclonal sera.
