EMD-28029
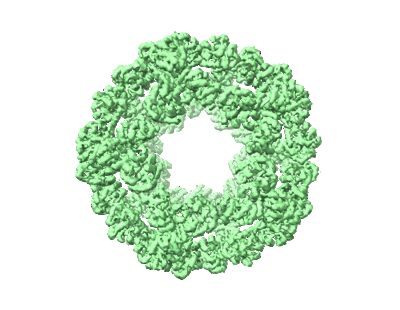
I3-01 map refined in C1
EMD-28029
Single-particle4.2 Å
 Deposition: 03/09/2022
Deposition: 03/09/2022Map released: 12/07/2023
Last modified: 12/07/2023
Improved interface packing and design opportunities revealed by CryoEM analysis of a designed protein nanocage.
Abstract:
Symmetric protein assemblies play important roles in nature which makes them an attractive target for engineering. De novo symmetric protein complexes can be created through computational protein design to tailor their properties from first principles, and recently several protein nanocages have been created by bringing together protein components through hydrophobic interactions. Accurate experimental structures of newly-developed proteins are essential to validate their design, improve assembly stability, and tailor downstream applications. We describe the CryoEM structure of the nanocage I3-01, at an overall resolution of 3.5 Å. I3-01, comprising 60 aldolase subunits arranged with icosahedral symmetry, has resisted high-resolution characterization. Some key differences between the refined structure and the original design are identified, such as improved packing of hydrophobic sidechains, providing insight to the resistance of I3-01 to high-resolution averaging. Based on our analysis, we suggest factors important in the design and structural processing of new assemblies.
Symmetric protein assemblies play important roles in nature which makes them an attractive target for engineering. De novo symmetric protein complexes can be created through computational protein design to tailor their properties from first principles, and recently several protein nanocages have been created by bringing together protein components through hydrophobic interactions. Accurate experimental structures of newly-developed proteins are essential to validate their design, improve assembly stability, and tailor downstream applications. We describe the CryoEM structure of the nanocage I3-01, at an overall resolution of 3.5 Å. I3-01, comprising 60 aldolase subunits arranged with icosahedral symmetry, has resisted high-resolution characterization. Some key differences between the refined structure and the original design are identified, such as improved packing of hydrophobic sidechains, providing insight to the resistance of I3-01 to high-resolution averaging. Based on our analysis, we suggest factors important in the design and structural processing of new assemblies.

