EMD-41207
Cryo-electron tomography reconstruction of mammalian 80S ribosome from EMPIAR-10064 in the translocation intermediates (TI) state with eEF2 cofactor and closed L1 by constrained classification.
EMD-41207
Subtomogram averaging10.0 Å
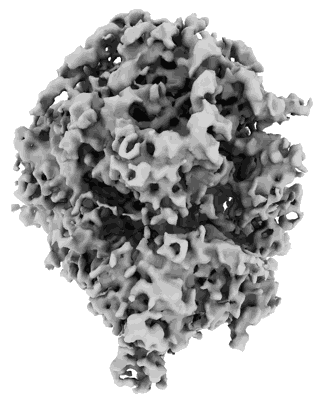 Deposition: 07/07/2023
Deposition: 07/07/2023Map released: 25/10/2023
Last modified: 20/12/2023
Sample Organism:
Oryctolagus cuniculus
Sample: Mammalian 80S ribosome in the translocation intermediates (TI) state with L1 stalk closed
Deposition Authors: Zhou Y ,
Liu HF
,
Liu HF  ,
Bartesaghi A
,
Bartesaghi A 
Sample: Mammalian 80S ribosome in the translocation intermediates (TI) state with L1 stalk closed
Deposition Authors: Zhou Y
 ,
Liu HF
,
Liu HF  ,
Bartesaghi A
,
Bartesaghi A 
nextPYP: a comprehensive and scalable platform for characterizing protein variability in situ using single-particle cryo-electron tomography.
Liu HF  ,
Zhou Y
,
Zhou Y  ,
Huang Q
,
Huang Q  ,
Piland J
,
Piland J  ,
Jin W,
Mandel J,
Du X
,
Jin W,
Mandel J,
Du X  ,
Martin J,
Bartesaghi A
,
Martin J,
Bartesaghi A 
(2023) Nat Methods , 20 , 1909 - 1919
 ,
Zhou Y
,
Zhou Y  ,
Huang Q
,
Huang Q  ,
Piland J
,
Piland J  ,
Jin W,
Mandel J,
Du X
,
Jin W,
Mandel J,
Du X  ,
Martin J,
Bartesaghi A
,
Martin J,
Bartesaghi A 
(2023) Nat Methods , 20 , 1909 - 1919
Abstract:
Single-particle cryo-electron tomography is an emerging technique capable of determining the structure of proteins imaged within the native context of cells at molecular resolution. While high-throughput techniques for sample preparation and tilt-series acquisition are beginning to provide sufficient data to allow structural studies of proteins at physiological concentrations, the complex data analysis pipeline and the demanding storage and computational requirements pose major barriers for the development and broader adoption of this technology. Here, we present a scalable, end-to-end framework for single-particle cryo-electron tomography data analysis from on-the-fly pre-processing of tilt series to high-resolution refinement and classification, which allows efficient analysis and visualization of datasets with hundreds of tilt series and hundreds of thousands of particles. We validate our approach using in vitro and cellular datasets, demonstrating its effectiveness at achieving high-resolution and revealing conformational heterogeneity in situ. The framework is made available through an intuitive and easy-to-use computer application, nextPYP ( http://nextpyp.app ).
Single-particle cryo-electron tomography is an emerging technique capable of determining the structure of proteins imaged within the native context of cells at molecular resolution. While high-throughput techniques for sample preparation and tilt-series acquisition are beginning to provide sufficient data to allow structural studies of proteins at physiological concentrations, the complex data analysis pipeline and the demanding storage and computational requirements pose major barriers for the development and broader adoption of this technology. Here, we present a scalable, end-to-end framework for single-particle cryo-electron tomography data analysis from on-the-fly pre-processing of tilt series to high-resolution refinement and classification, which allows efficient analysis and visualization of datasets with hundreds of tilt series and hundreds of thousands of particles. We validate our approach using in vitro and cellular datasets, demonstrating its effectiveness at achieving high-resolution and revealing conformational heterogeneity in situ. The framework is made available through an intuitive and easy-to-use computer application, nextPYP ( http://nextpyp.app ).
