EMD-0441
Conformational switches control early maturation of the eukaryotic small ribosomal subunit
EMD-0441
Single-particle4.3 Å
 Deposition: 13/12/2018
Deposition: 13/12/2018Map released: 19/06/2019
Last modified: 20/03/2024
Sample Organism:
Saccharomyces cerevisiae BY4741
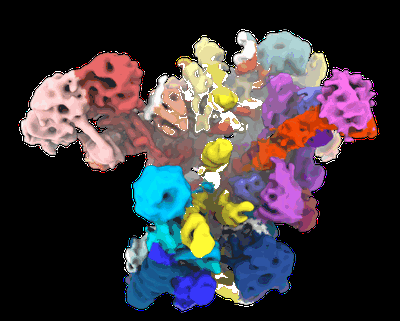
Sample: 5' ETS particle
Fitted models: 6nd4 (Avg. Q-score: 0.26)
Deposition Authors: Hunziker M ,
Barandun J
,
Barandun J 
Sample: 5' ETS particle
Fitted models: 6nd4 (Avg. Q-score: 0.26)
Deposition Authors: Hunziker M
 ,
Barandun J
,
Barandun J 
Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Abstract:
Eukaryotic ribosome biogenesis is initiated with the transcription of pre-ribosomal RNA at the 5' external transcribed spacer, which directs the early association of assembly factors but is absent from the mature ribosome. The subsequent co-transcriptional association of ribosome assembly factors with pre-ribosomal RNA results in the formation of the small subunit processome. Here we show that stable rRNA domains of the small ribosomal subunit can independently recruit their own biogenesis factors in vivo. The final assembly and compaction of the small subunit processome requires the presence of the 5' external transcribed spacer RNA and all ribosomal RNA domains. Additionally, our cryo-electron microscopy structure of the earliest nucleolar pre-ribosomal assembly - the 5' external transcribed spacer ribonucleoprotein - provides a mechanism for how conformational changes in multi-protein complexes can be employed to regulate the accessibility of binding sites and therefore define the chronology of maturation events during early stages of ribosome assembly.
Eukaryotic ribosome biogenesis is initiated with the transcription of pre-ribosomal RNA at the 5' external transcribed spacer, which directs the early association of assembly factors but is absent from the mature ribosome. The subsequent co-transcriptional association of ribosome assembly factors with pre-ribosomal RNA results in the formation of the small subunit processome. Here we show that stable rRNA domains of the small ribosomal subunit can independently recruit their own biogenesis factors in vivo. The final assembly and compaction of the small subunit processome requires the presence of the 5' external transcribed spacer RNA and all ribosomal RNA domains. Additionally, our cryo-electron microscopy structure of the earliest nucleolar pre-ribosomal assembly - the 5' external transcribed spacer ribonucleoprotein - provides a mechanism for how conformational changes in multi-protein complexes can be employed to regulate the accessibility of binding sites and therefore define the chronology of maturation events during early stages of ribosome assembly.
