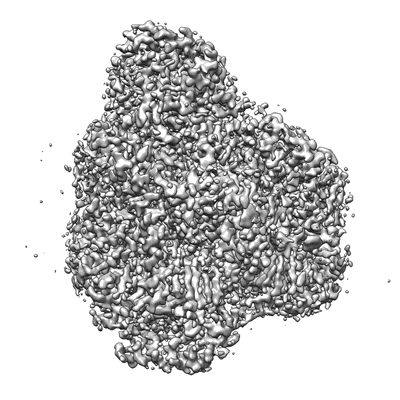
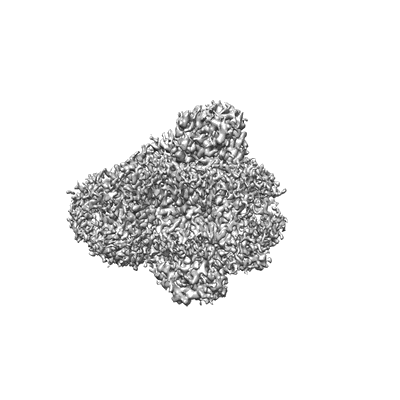
Cryo-EM structure of SoxS-dependent transcription activation complex with micF promoter DNA
Resolution: 4.55 Å
EM Method: Single-particle
Fitted PDBs: 7w5w
Q-score: 0.332
Shi J, Wang L, Wen A, Wang F, Zhang Y, Yu L, Li F, Jin Y, Feng Z, Li J, Yang Y, Gao F, Zhang Y, Feng Y, Wang S, Zhao W, Lin W
Nucleic Acids Res (2022) 50 pp. 11359-11373 [ Pubmed: 36243985 DOI: doi:10.1093/nar/gkac898 ]
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli K-12)
- Dna (Complex)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli K-12)
- Micf promoter dna reverse strand (21 kDa, DNA from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- Soxs-dependent transcription activation complex with micf promoter dna (Complex from Escherichia coli K-12)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunits (Complex from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli K-12)
- Regulatory protein soxs (12 kDa, Protein from Escherichia coli K-12)
- Micf promoter dna forward strand (21 kDa, DNA from Escherichia coli)
Cryo-EM structure of SoxS-dependent transcription activation complex with zwf promoter DNA
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7w5x
Q-score: 0.47
Shi J, Wang L, Wen A, Wang F, Zhang Y, Yu L, Li F, Jin Y, Feng Z, Li J, Yang Y, Gao F, Zhang Y, Feng Y, Wang S, Zhao W, Lin W
Nucleic Acids Res (2022) 50 pp. 11359-11373 [ Pubmed: 36243985 DOI: doi:10.1093/nar/gkac898 ]
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli K-12)
- Dna (Complex)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli K-12)
- Regulatory protein soxs (12 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli K-12)
- Zwf promoter dna reverse strand (23 kDa, DNA from Escherichia coli)
- Soxs-dependent transcription activation complex with zwf promoter dna (Complex from Escherichia coli K-12)
- Dna-directed rna polymerase subunits (Complex from Escherichia coli)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli K-12)
- Zwf promoter dna forward strand (23 kDa, DNA from Escherichia coli)
Cryo-EM structure of SoxS-dependent transcription activation complex with fpr promoter DNA
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 7w5y
Q-score: 0.369
Shi J, Wang L, Wen A, Wang F, Zhang Y, Yu L, Li F, Jin Y, Feng Z, Li J, Yang Y, Gao F, Zhang Y, Feng Y, Wang S, Zhao W, Lin W
Nucleic Acids Res (2022) 50 pp. 11359-11373 [ Pubmed: 36243985 DOI: doi:10.1093/nar/gkac898 ]
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli K-12)
- Dna (Complex)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli K-12)
- Soxs-dependent transcription activation complex with fpr promoter dna (Complex from Escherichia coli K-12)
- Regulatory protein soxs (12 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli K-12)
- Fpr promoter dna reverse strand (26 kDa, DNA from Escherichia coli)
- Dna-directed rna polymerase subunits (Complex from Escherichia coli)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli K-12)
- Fpr promoter dna forward strand (26 kDa, DNA from Escherichia coli)
Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation
Resolution: 4.57 Å
EM Method: Single-particle
Fitted PDBs: 7vwy
Q-score: 0.359
Shi J, Wang F, Li F, Wang L, Xiong Y, Wen A, Jin Y, Jin S, Gao F, Feng Z, Li J, Zhang Y, Shang Z, Wang S, Feng Y, Lin W
Nucleic Acids Res (2022) 50 pp. 5974-5987 [ DOI: doi:10.1093/nar/gkac433 Pubmed: 35641097 ]
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli K-12)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli K-12)
- Zinc ion (65 Da, Ligand)
- Right origin-binding protein (33 kDa, Protein from Escherichia coli K-12)
- Rob-dependent transcription activation complex (Complex from Escherichia coli)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli K-12)
- Micf promoter dna scaffold forward strand (21 kDa, DNA from Escherichia coli)
- Micf promoter dna scaffold reverse strand (21 kDa, DNA from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli K-12)
Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 7vwz
Q-score: 0.345
Shi J, Wang F, Li F, Wang L, Xiong Y, Wen A, Jin Y, Jin S, Gao F, Feng Z, Li J, Zhang Y, Shang Z, Wang S, Feng Y, Lin W
Nucleic Acids Res (2022) 50 pp. 5974-5987 [ DOI: doi:10.1093/nar/gkac433 Pubmed: 35641097 ]
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli K-12)
- Dna (Complex)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli K-12)
- Micf promoter dna reverse strand (21 kDa, DNA from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- Right origin-binding protein (33 kDa, Protein from Escherichia coli K-12)
- Rna polymerase subunits (Complex from Escherichia coli)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli K-12)
- Rob-dependent transcription activation complex (Complex from Escherichia coli K-12)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli K-12)
- Micf promoter dna forward strand (21 kDa, DNA from Escherichia coli)
E. coli cytochrome bo3 in MSP nanodisc
Resolution: 2.19 Å
EM Method: Single-particle
Fitted PDBs: 7n9z
Q-score: 0.734
Li J, Han L, Vallese F, Ding Z, Choi SK, Hong S, Luo Y, Liu B, Chan CK, Tajkhorshid E, Zhu J, Clarke O, Zhang K, Gennis R
PNAS (2021) 118 [ Pubmed: 34417297 DOI: doi:10.1073/pnas.2106750118 ]
- Zinc ion (65 Da, Ligand)
- Cytochrome o ubiquinol oxidase (22 kDa, Protein from Escherichia coli)
- Ubiquinone-8 (727 Da, Ligand)
- Cytochrome o ubiquinol oxidase, subunit i (74 kDa, Protein from Escherichia coli)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Cytochrome o ubiquinol oxidase, subunit iv (12 kDa, Protein from Escherichia coli)
- Heme o (838 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Cytochrome bo 3 ubiquinol oxidase (Complex from Escherichia coli)
- Ubiquinol oxidase subunit 2 (34 kDa, Protein from Escherichia coli)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Copper (ii) ion (63 Da, Ligand)
- Cardiolipin (1 kDa, Ligand)
- Water (18 Da, Ligand)
2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane
Resolution: 2.55 Å
EM Method: Single-particle
Fitted PDBs: 7cub
Q-score: 0.645
Li J, Han L, Vallese F, Ding Z, Choi SK, Hong S, Luo Y, Liu B, Chan CK, Tajkhorshid E, Zhu J, Clarke O, Zhang K, Gennis R
PNAS (2021) 118 [ Pubmed: 34417297 DOI: doi:10.1073/pnas.2106750118 ]
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase (140 kDa, Complex from Escherichia coli)
- Ubiquinone-8 (727 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 4 (12 kDa, Protein from Escherichia coli)
- Heme o (838 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 1 (74 kDa, Protein from Escherichia coli)
- Copper (ii) ion (63 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 3 (22 kDa, Protein from Escherichia coli)
- Cytochrome bo(3) ubiquinol oxidase subunit 2 (32 kDa, Protein from Escherichia coli)
- Water (18 Da, Ligand)
2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 7cuq
Q-score: 0.597
Li J, Han L, Vallese F, Ding Z, Choi SK, Hong S, Luo Y, Liu B, Chan CK, Tajkhorshid E, Zhu J, Clarke O, Zhang K, Gennis R
PNAS (2021) 118 [ Pubmed: 34417297 DOI: doi:10.1073/pnas.2106750118 ]
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase (140 kDa, Complex from Escherichia coli)
- Ubiquinone-8 (727 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 4 (12 kDa, Protein from Escherichia coli)
- Heme o (838 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 1 (74 kDa, Protein from Escherichia coli)
- Copper (ii) ion (63 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 3 (22 kDa, Protein from Escherichia coli)
- Cytochrome bo(3) ubiquinol oxidase subunit 2 (32 kDa, Protein from Escherichia coli)
Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli
Resolution: 2.63 Å
EM Method: Single-particle
Fitted PDBs: 7cuw
Q-score: 0.63
Li J, Han L, Vallese F, Ding Z, Choi SK, Hong S, Luo Y, Liu B, Chan CK, Tajkhorshid E, Zhu J, Clarke O, Zhang K, Gennis R
PNAS (2021) 118 [ Pubmed: 34417297 DOI: doi:10.1073/pnas.2106750118 ]
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase (140 kDa, Complex from Escherichia coli)
- Ubiquinone-8 (727 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 4 (12 kDa, Protein from Escherichia coli)
- Heme o (838 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 1 (74 kDa, Protein from Escherichia coli)
- Copper (ii) ion (63 Da, Ligand)
- Cytochrome bo(3) ubiquinol oxidase subunit 3 (22 kDa, Protein from Escherichia coli)
- Cytochrome bo(3) ubiquinol oxidase subunit 2 (32 kDa, Protein from Escherichia coli)
Structure of the CS1 pilus of enterotoxigenic Escherichia coli reveals structural polymorphism
Galkin VE, Kolappan S, Ng D, Zong Z, Li J, Yu X, Egelman EH, Craig L
J Bacteriol (2013) 195 pp. 1360-1370 [ Pubmed: 23175654 DOI: doi:10.1128/JB.01989-12 ]
- Cs1 pilus (Sample)
- Cooa (15 kDa, Cellular component from Escherichia coli)