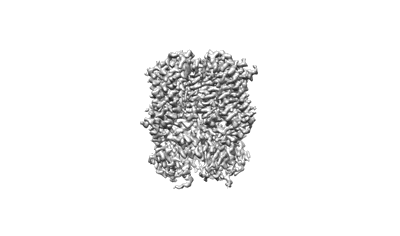
Merbecovirus MOW15-22 Spike glycoprotein RBD bound to the P. davyi ACE2
Ma CB, Liu C, Park YJ, Tang J, Chen J, Xiong Q, Lee J, Stewart C, Asarnow D, Brown J, Tortorici MA, Yang X, Sun YH, Chen YM, Yu X, Si JY, Liu P, Tong F, Huang ML, Li J, Shi ZL, Deng Z, Veesler D, Yan H
Cell (2025) [ Pubmed: 39922191 DOI: doi:10.1016/j.cell.2024.12.031 ]
- Mow15-22 rbd (34 kDa, Protein from Merbecovirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Pteronotus davyi ace2 ectodomain (89 kDa, Protein from Pteronotus davyi)
- Mow15-22 rbd bound to the p. davyi ace2 ectodomain (Complex from Sarbecovirus)
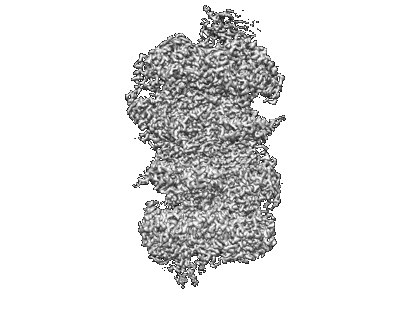
Merbecovirus PnNL2018B Spike glycoprotein RBD bound to the P. Nathusii ACE2
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 9dak
Q-score: 0.632
Ma CB, Liu C, Park YJ, Tang J, Chen J, Xiong Q, Lee J, Stewart C, Asarnow D, Brown J, Tortorici MA, Yang X, Sun YH, Chen YM, Yu X, Si JY, Liu P, Tong F, Huang ML, Li J, Shi ZL, Deng Z, Veesler D, Yan H
Cell (2025) [ DOI: doi:10.1016/j.cell.2024.12.031 Pubmed: 39922191 ]
- Ace2 (102 kDa, Protein from Pipistrellus nathusii)
- Water (18 Da, Ligand)
- Merbecovirus pnnl2018b spike glycoprotein rbd bound to the p. nathusii ace2 (Complex from Merbecovirus)
- Spike glycoprotein receptor binding domain (34 kDa, Protein from Merbecovirus)
- Zinc ion (65 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 9c44
Q-score: 0.535
Liu P, Huang ML, Guo H, McCallum M, Si JY, Chen YM, Wang CL, Yu X, Shi LL, Xiong Q, Ma CB, Bowen JE, Tong F, Liu C, Sun YH, Yang X, Chen J, Guo M, Li J, Corti D, Veesler D, Shi ZL, Yan H
Nature (2024) 635 pp. 978-986 [ DOI: doi:10.1038/s41586-024-08121-5 Pubmed: 39478224 ]
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- S2l20 fab light chain variable region (11 kDa, Protein from Homo sapiens)
- Sars-cov-2 s bound by s2l20 fab (Complex from Homo sapiens)
- S2l20 fab heavy chain variable (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
The structure of HKU1-B S protein with bsAb1
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8yww
Q-score: 0.291
Sun H, Xia L, Li J, Zhang Y, Zhang G, Huang P, Wang X, Cui Y, Fang T, Fan P, Zhou Q, Chi X, Yu C
Emerg Microbes Infect (2024) 13 pp. 2373307-2373307 [ DOI: doi:10.1080/22221751.2024.2373307 Pubmed: 38953857 ]
- Spike glycoprotein (134 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- The structure of the bsab1-omicron ba.5 s complex (Complex from Severe acute respiratory syndrome coronavirus 2)
- H4b6 light chain (23 kDa, Protein from Homo sapiens)
- The scfv of h4d12 (26 kDa, Protein from Homo sapiens)
- H4b6 heavy chain (23 kDa, Protein from Homo sapiens)
Cryo-EM structure of an anti-phage defense complex
Resolution: 2.87 Å
EM Method: Single-particle
Fitted PDBs: 8xav
Q-score: 0.535
An Q, Wang Y, Tian Z, Han J, Li J, Liao F, Yu F, Zhao H, Wen Y, Zhang H, Deng Z
Cell Res (2024) 34 pp. 545-555 [ Pubmed: 38834762 DOI: doi:10.1038/s41422-024-00981-w ]
- Duf4297 (46 kDa, Protein from Escherichia coli)
- Duf4297-hera (Complex from Escherichia coli)
- Atp-binding protein (64 kDa, Protein from Escherichia coli)
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 1
Resolution: 2.73 Å
EM Method: Single-particle
Fitted PDBs: 8xaw
Q-score: 0.553
An Q, Wang Y, Tian Z, Han J, Li J, Liao F, Yu F, Zhao H, Wen Y, Zhang H, Deng Z
Cell Res (2024) 34 pp. 545-555 [ Pubmed: 38834762 DOI: doi:10.1038/s41422-024-00981-w ]
- Magnesium ion (24 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- S20dna1 (18 kDa, DNA from Escherichia coli)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- S20dna2 (18 kDa, DNA from Escherichia coli)
- Atp-binding protein (64 kDa, Protein from Escherichia coli)
- Duf4297 (46 kDa, Protein from Escherichia coli)
- Duf4297-hera-dna-state2 (Complex from Escherichia coli)
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 2
Resolution: 2.92 Å
EM Method: Single-particle
Fitted PDBs: 8xax
Q-score: 0.516
An Q, Wang Y, Tian Z, Han J, Li J, Liao F, Yu F, Zhao H, Wen Y, Zhang H, Deng Z
Cell Res (2024) 34 pp. 545-555 [ Pubmed: 38834762 DOI: doi:10.1038/s41422-024-00981-w ]
- Magnesium ion (24 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- Duf4297-hera-dna state1 (Complex from Escherichia coli)
- S20dna1 (18 kDa, DNA from Escherichia coli)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- S20dna2 (18 kDa, DNA from Escherichia coli)
- Atp-binding protein (64 kDa, Protein from Escherichia coli)
- Duf4297 (46 kDa, Protein from Escherichia coli)
Cryo-EM structure of an anti-phage defense complex bound to ATPrS and DNA
Resolution: 2.81 Å
EM Method: Single-particle
Fitted PDBs: 8xay
Q-score: 0.55
An Q, Wang Y, Tian Z, Han J, Li J, Liao F, Yu F, Zhao H, Wen Y, Zhang H, Deng Z
Cell Res (2024) 34 pp. 545-555 [ Pubmed: 38834762 DOI: doi:10.1038/s41422-024-00981-w ]
- Magnesium ion (24 Da, Ligand)
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- S20dna3 (18 kDa, Macromolecule from Escherichia coli)
- Atp-binding protein (64 kDa, Protein from Escherichia coli)
- Duf4297 (46 kDa, Protein from Escherichia coli)
- Duf4297-hera (Complex from Escherichia coli)
- S20dna4 (18 kDa, DNA from Escherichia coli)
Cryo-EM structure of human HCN3 channel in apo state
Resolution: 2.72 Å
EM Method: Single-particle
Fitted PDBs: 8inz
Q-score: 0.526
Yu B, Lu Q, Li J, Cheng X, Hu H, Li Y, Che T, Hua Y, Jiang H, Zhang Y, Xian C, Yang T, Fu Y, Chen Y, Nan W, McCormick PJ, Xiong B, Duan J, Zeng B, Li Y, Fu Y, Zhang J
J Biol Chem (2024) 300 pp. 107288-107288 [ Pubmed: 38636662 DOI: doi:10.1016/j.jbc.2024.107288 ]
- 4-[[(2~{s},4~{a}~{r},6~{s},8~{a}~{s})-6-[(4~{s},5~{r})-4-[(2~{s})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{h}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid (492 Da, Ligand)
- Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (86 kDa, Protein from Homo sapiens)
- Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Complex from Homo sapiens)
Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA)
Resolution: 2.59 Å
EM Method: Single-particle
Fitted PDBs: 7w1y
Q-score: 0.595
Li J, Dong J, Wang W, Yu D, Fan X, Hui YC, Lee CSK, Lam WH, Alary N, Yang Y, Zhang Y, Zhao Q, Chen CL, Tye BK, Dang S, Zhai Y
Cell (2023) 186 pp. 98-111.e21 [ Pubmed: 36608662 DOI: doi:10.1016/j.cell.2022.12.008 ]
- Dna replication licensing factor mcm5 (82 kDa, Protein from Homo sapiens)
- Dna (49-mer) (14 kDa, DNA from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Isoform 2 of dna replication licensing factor mcm3 (96 kDa, Protein from Homo sapiens)
- Dna replication licensing factor mcm6 (93 kDa, Protein from Homo sapiens)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dna replication licensing factor mcm4 (96 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Human mini-chromosome maintenance complex (1 MDa, Complex from Homo sapiens)
- Dna replication licensing factor mcm7 (81 kDa, Protein from Homo sapiens)
- Dna (49-mer) (15 kDa, DNA from Homo sapiens)
- Dna replication licensing factor mcm2 (102 kDa, Protein from Homo sapiens)
- Dna (Complex)
- Dna replication licensing factor mcm 2/3/4/5/6/7 (Complex from Homo sapiens)