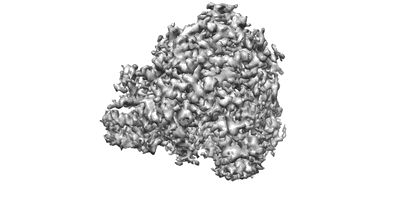
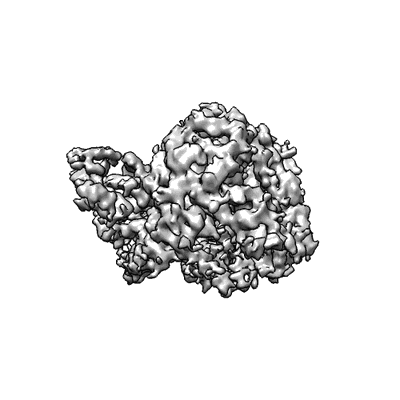CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8k4s
Q-score: 0.521
Yang J, Zhao T, Fan J, Zou H, Lan G, Guo F, Shi Y, Ke H, Yu H, Yue Z, Wang X, Bai Y, Li S, Liu Y, Wang X, Chen Y, Li Y, Lei X
Cell (2024) 187 pp. 7164-7182.e18 [ DOI: doi:10.1016/j.cell.2024.10.001 Pubmed: 39476841 ]
- (4~{r})-4-[(3~{r},5~{r},8~{r},9~{s},10~{s},12~{s},13~{r},14~{s},17~{r})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{h}-cyclopenta[a]phenanthren-17-yl]pentanoic acid (472 Da, Ligand)
- Soluble cytochrome b562,mas-related g-protein coupled receptor member x4,green fluorescent protein (79 kDa, Protein from Aequorea victoria)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (8 kDa, Protein from Homo sapiens)
- Complex of mrgprx4 with gq and agonist dca-3p (Complex from Homo sapiens)
- Scfv16 (26 kDa, Protein from Mus musculus)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Homo sapiens)
- Gs-mini-gq chimera (28 kDa, Protein from Homo sapiens)
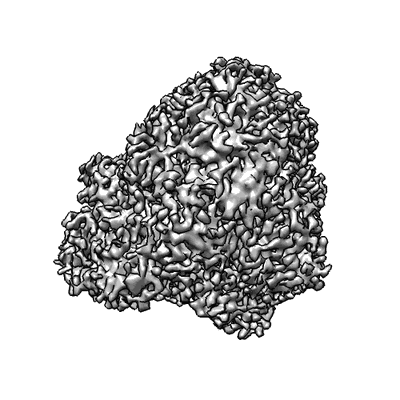
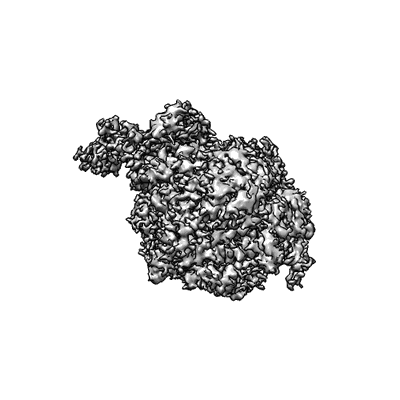
CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P, local
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8kex
Q-score: 0.416
Yang J, Zhao T, Fan J, Zou H, Lan G, Guo F, Shi Y, Ke H, Yu H, Yue Z, Wang X, Bai Y, Li S, Liu Y, Wang X, Chen Y, Li Y, Lei X
Cell (2024) 187 pp. 7164-7182.e18 [ DOI: doi:10.1016/j.cell.2024.10.001 Pubmed: 39476841 ]
- (4~{r})-4-[(3~{r},5~{r},8~{r},9~{s},10~{s},12~{s},13~{r},14~{s},17~{r})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{h}-cyclopenta[a]phenanthren-17-yl]pentanoic acid (472 Da, Ligand)
- Soluble cytochrome b562,mas-related g-protein coupled receptor member x4,green fluorescent protein (79 kDa, Protein from Aequorea victoria)
- Cryo-em structure of mrgprx4 complex with agonist dca-3p (Complex from Homo sapiens)
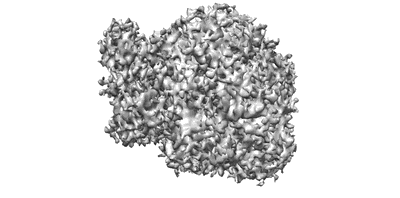
Cryo-EM structures of Alphacoronavirus spike glycoprotein
Resolution: 3.21 Å
EM Method: Single-particle
Fitted PDBs: 7cyc
Q-score: 0.424
Song X, Shi Y, Ding W, Niu T, Sun L, Tan Y, Chen Y, Shi J, Xiong Q, Huang X, Xiao S, Zhu Y, Cheng C, Fu ZF, Liu ZJ, Peng G
Nat Commun (2021) 12 pp. 141-141 [ DOI: doi:10.1038/s41467-020-20401-y Pubmed: 33420048 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Hcov-229e spike trimer (Complex from Human coronavirus 229E)
- Spike glycoprotein (122 kDa, Protein from Human coronavirus 229E)
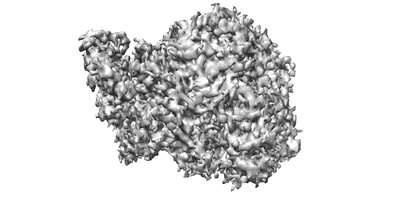
Cryo-EM structures of Alphacoronavirus spike glycoprotein
Resolution: 3.55 Å
EM Method: Single-particle
Fitted PDBs: 7cyd
Q-score: 0.361
Song X, Shi Y, Ding W, Niu T, Sun L, Tan Y, Chen Y, Shi J, Xiong Q, Huang X, Xiao S, Zhu Y, Cheng C, Fu ZF, Liu ZJ, Peng G
Nat Commun (2021) 12 pp. 141-141 [ DOI: doi:10.1038/s41467-020-20401-y Pubmed: 33420048 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Hcov-229e spike trimer (Complex from Human coronavirus 229E)
- Spike glycoprotein (122 kDa, Protein from Human coronavirus 229E)
structure of influenza D virus apo polymerase
Resolution: 4.6 Å
EM Method: Single-particle
Fitted PDBs: 6kv5
Q-score: 0.213
Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, Wang P, Yan J, Zhang B, Qi J, Deng T, Gao GF, Shi Y
Nat Microbiol (2019) [ DOI: doi:10.1038/s41564-019-0487-5 Pubmed: 31209309 ]
- Rna-directed rna polymerase catalytic subunit (86 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Polymerase 3 (83 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Influenza d virus polymerase (270 kDa, Complex from Thogotovirus)
- Polymerase pb2 (88 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1)
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 6kuk
Q-score: 0.403
Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, Wang P, Yan J, Zhang B, Qi J, Deng T, Gao GF, Shi Y
Nat Microbiol (2019) [ DOI: doi:10.1038/s41564-019-0487-5 Pubmed: 31209309 ]
- Rna-directed rna polymerase catalytic subunit (86 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Polymerase 3 (83 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Rna (5'-r(p*cp*up*cp*cp*up*gp*cp*up*up*ap*up*gp*cp*u)-3') (4 kDa, RNA from synthetic construct)
- Polymerase pb2 (88 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Rna (5'-r(p*ap*gp*cp*ap*gp*up*ap*gp*cp*ap*ap*gp*gp*ap*g)-3') (4 kDa, RNA from synthetic construct)
- Influenza d virus polymerase bound to vrna promoter (290 kDa, Complex from Influenza D virus)
Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2)
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 6kup
Q-score: 0.326
Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, Wang P, Yan J, Zhang B, Qi J, Deng T, Gao GF, Shi Y
Nat Microbiol (2019) [ DOI: doi:10.1038/s41564-019-0487-5 Pubmed: 31209309 ]
- Rna-directed rna polymerase catalytic subunit (86 kDa, Protein from Influenza D virus)
- Influenza d virus polymerase (270 kDa, Complex from Influenza D virus)
- Polymerase 3 (83 kDa, Protein from Influenza D virus)
- Rna (5'-r(p*cp*up*cp*cp*up*gp*cp*up*up*ap*up*gp*cp*u)-3') (4 kDa, RNA from synthetic construct)
- Polymerase pb2 (88 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Rna (5'-r(p*ap*gp*cp*ap*gp*up*ap*gp*cp*ap*ap*gp*gp*ap*g)-3') (4 kDa, RNA from synthetic construct)
structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2)
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 6kut
Q-score: 0.358
Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, Wang P, Yan J, Zhang B, Qi J, Deng T, Gao GF, Shi Y
Nat Microbiol (2019) [ DOI: doi:10.1038/s41564-019-0487-5 Pubmed: 31209309 ]
- Rna-directed rna polymerase catalytic subunit (86 kDa, Protein from Influenza D virus)
- Polymerase 3 (83 kDa, Protein from Influenza D virus)
- Rna (5'-r(p*cp*up*cp*cp*up*gp*cp*up*up*ap*up*gp*cp*u)-3') (4 kDa, RNA from synthetic construct)
- Polymerase pb2 (88 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Rna (5'-r(p*ap*gp*cp*ap*gp*up*ap*gp*cp*ap*ap*gp*gp*ap*g)-3') (4 kDa, RNA from synthetic construct)
- Influenza d virus polymerase bound to vrna promoter in mode b conformation (class b2) (270 kDa, Complex from Thogotovirus)
Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1)
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6kur
Q-score: 0.42
Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, Wang P, Yan J, Zhang B, Qi J, Deng T, Gao GF, Shi Y
Nat Microbiol (2019) [ DOI: doi:10.1038/s41564-019-0487-5 Pubmed: 31209309 ]
- Rna-directed rna polymerase catalytic subunit (86 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Influenza d virus polymerase bound to vrna promoter (270 kDa, Complex from Influenza D virus)
- Polymerase 3 (83 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Polymerase pb2 (29 kDa, Protein from Influenza D virus (D/swine/Oklahoma/1334/2011))
- Rna (5'-r(p*cp*up*cp*cp*up*gp*cp*up*up*ap*up*gp*cp*u)-3') (4 kDa, RNA from synthetic construct)
- Rna (5'-r(p*ap*gp*cp*ap*gp*up*ap*gp*cp*ap*ap*gp*gp*ap*g)-3') (4 kDa, RNA from synthetic construct)
Structure of influenza D virus polymerase bound to cRNA promoter in class 1
Resolution: 3.44 Å
EM Method: Single-particle
Fitted PDBs: 6kuj
Q-score: 0.481
Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, Wang P, Yan J, Zhang B, Qi J, Deng T, Gao GF, Shi Y
Nat Microbiol (2019) [ DOI: doi:10.1038/s41564-019-0487-5 Pubmed: 31209309 ]
- Rna-directed rna polymerase catalytic subunit (86 kDa, Protein from Influenza D virus)
- Rna (5'-r(p*up*cp*cp*up*up*gp*cp*up*ap*cp*up*gp*cp*u)-3') (4 kDa, RNA from synthetic construct)
- Polymerase 3 (83 kDa, Protein from Influenza D virus)
- Influenza d virus polymerase bound to crna promoter in mode a conformation (270 kDa, Complex from Influenza D virus)
- Polymerase pb2 (88 kDa, Protein from Influenza D virus)
- Rna (5'-r(p*ap*gp*cp*ap*up*ap*ap*gp*cp*ap*gp*gp*ap*gp*a)-3') (4 kDa, RNA from synthetic construct)