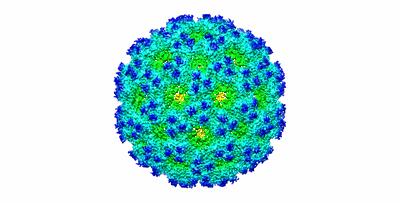
Cryo-EM structure of human SARM1 TIR domain in complex with 1AD
Shi Y, Kerry PS, Nanson JD, Bosanac T, Sasaki Y, Krauss R, Saikot FK, Adams SE, Mosaiab T, Masic V, Mao X, Rose F, Vasquez E, Furrer M, Cunnea K, Brearley A, Gu W, Luo Z, Brillault L, Landsberg MJ, DiAntonio A, Kobe B, Milbrandt J, Hughes RO, Ve T
Mol Cell (2022) 82 pp. 1643-1659.e10 [ Pubmed: 35334231 DOI: doi:10.1016/j.molcel.2022.03.007 ]
Double nuclear outer ring from the isolated yeast NPC
Resolution: 11.6 Å
EM Method: Single-particle
Fitted PDBs: 7n84
Q-score: 0.063
Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, Salzberg D, Song K, Xu C, Gumbart JC, Suslov S, Unruh J, Jaspersen SL, Chait BT, Sali A, Fernandez-Martinez J, Ludtke SJ, Villa E, Rout MP
Cell (2022) 185 pp. 361-378.e25 [ Pubmed: 34982960 DOI: doi:10.1016/j.cell.2021.12.015 ]
- Nucleoporin seh1 (39 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin 145c (81 kDa, Protein from Saccharomyces cerevisiae)
- Yeast double outer ring (10 MDa, Complex from Saccharomyces cerevisiae)
- Nucleoporin nup120 (120 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup84 (83 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup188 (188 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup133 (133 kDa, Protein from Saccharomyces cerevisiae)
- Unknown (5 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup85 (84 kDa, Protein from Saccharomyces cerevisiae)
- Protein transport protein sec13 (33 kDa, Protein from Saccharomyces cerevisiae)
Cryo-EM map of yeast 26S proteasome in M2 state derived from Arctica dataset
Luan B, Huang X, Wu J, Mei Z, Wang Y, Xue X, Yan C, Wang J, Finley DJ, Shi Y, Wang F
PNAS (2016) 113 pp. 2642-2647 [ DOI: doi:10.1073/pnas.1601561113 Pubmed: 26929360 ]
- Yeast 26s proteasome in m2 state (2 MDa, Sample)
- 26s proteasome (2 MDa, Protein from Saccharomyces cerevisiae)
Cryo-EM map of yeast 26S proteasome in M2 state with RP mask derived from Arctica dataset
Luan B, Huang X, Wu J, Mei Z, Wang Y, Xue X, Yan C, Wang J, Finley DJ, Shi Y, Wang F
PNAS (2016) 113 pp. 2642-2647 [ DOI: doi:10.1073/pnas.1601561113 Pubmed: 26929360 ]
- Rp of yeast 26s proteasome in m2 state (2 MDa, Sample)
- 19s rp of proteasome (2 MDa, Protein from Saccharomyces cerevisiae)
Resolution: 9.0 Å
EM Method: Single-particle
Fitted PDBs: 3j3t
Q-score: 0.077
Liu J, Mei Z, Li N, Qi Y, Xu Y, Shi Y, Wang F, Lei J, Gao N
J Biol Chem (2013) 288 pp. 17597-17608 [ Pubmed: 23595989 DOI: doi:10.1074/jbc.M113.458752 ]
- Meca-clpc(e280a) (600 kDa, Sample)
- Meca (Protein from Bacillus subtilis)
- Clpc (Protein from Bacillus subtilis)
Resolution: 11.0 Å
EM Method: Single-particle
Fitted PDBs: 3j3s
Q-score: 0.104
Liu J, Mei Z, Li N, Qi Y, Xu Y, Shi Y, Wang F, Lei J, Gao N
J Biol Chem (2013) 288 pp. 17597-17608 [ Pubmed: 23595989 DOI: doi:10.1074/jbc.M113.458752 ]
- Meca-clpc(e618a) (600 kDa, Sample)
- Meca (Protein from Bacillus subtilis)
- Clpc (Protein from Bacillus subtilis)
Resolution: 10.0 Å
EM Method: Single-particle
Fitted PDBs: 3j3u
Q-score: 0.086
Liu J, Mei Z, Li N, Qi Y, Xu Y, Shi Y, Wang F, Lei J, Gao N
J Biol Chem (2013) 288 pp. 17597-17608 [ Pubmed: 23595989 DOI: doi:10.1074/jbc.M113.458752 ]
- Meca-clpc (e280a,e618a) with atp (600 kDa, Sample)
- Meca (Protein from Bacillus subtilis)
- Clpc (Protein from Bacillus subtilis)
Resolution: 9.4 Å
EM Method: Single-particle
Fitted PDBs: 3j3r
Q-score: 0.079
Liu J, Mei Z, Li N, Qi Y, Xu Y, Shi Y, Wang F, Lei J, Gao N
J Biol Chem (2013) 288 pp. 17597-17608 [ Pubmed: 23595989 DOI: doi:10.1074/jbc.M113.458752 ]
- Meca (Protein from Bacillus subtilis)
- Clpc (Protein from Bacillus subtilis)
- Meca-clpc(e280a,e618a)with adp (600 kDa, Sample)