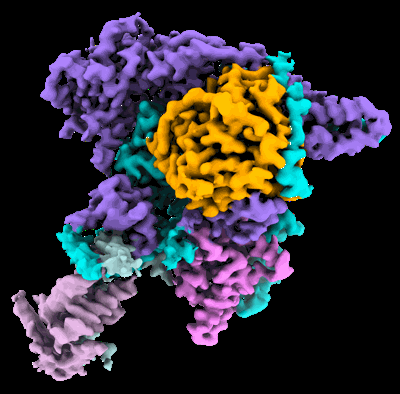
Cryo-EM structure of the histone deacetylase complex Rpd3S
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8hxx
Q-score: 0.453
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
- Zinc ion (65 Da, Ligand)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with histone h3 tail (Complex from Saccharomyces cerevisiae)
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
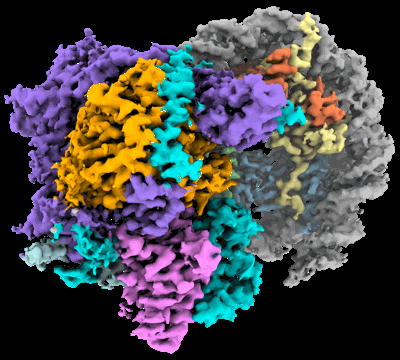
Cryo-EM structure of the histone deacetylase complex Rpd3S in complex with nucleosome
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8hxy
Q-score: 0.44
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
- Rpd3s histone deacetylase in complex with nucleosome (680 kDa, Complex from Saccharomyces cerevisiae)
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Transcriptional regulatory protein rco1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
Cryo-EM structure of Eaf3 CHD in complex with nucleosome
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8hxz
Q-score: 0.326
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Eaf3 chd in complex with nucleosome (150 kDa, Complex from Saccharomyces)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
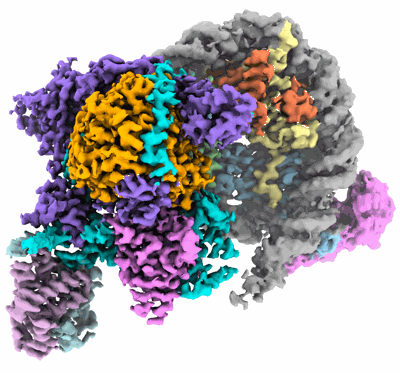
Composite cryo-EM structure of the histone deacetylase complex Rpd3S in complex with nucleosome
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8hy0
Q-score: 0.463
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with nucleosome (800 kDa, Complex from Saccharomyces cerevisiae)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
Concensus map of the Rpd3S-nucleosome complex
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
Focused refinement map of nucleosome in the Rpd3S-nucleosome complex
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
Focused refinement map of Rpd3S in the Rpd3S-nucleosome complex
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
Focused refinement map of the tail of Rpd3S
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
Focused refinement map of the CHD of Eaf3 in the Rpd3S-nucleosome complex
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
Cryo-EM structure of the histone deacetylase complex Rpd3S in complex with di-nucleosome
Resolution: 7.6 Å
EM Method: Single-particle
Fitted PDBs: 8jho
Q-score: 0.067
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ DOI: doi:10.1038/s41594-023-01121-5 Pubmed: 37798513 ]
- Di-nucleosome template reverse (108 kDa, DNA from synthetic construct)
- The rpd3s hdac complex (Complex from Saccharomyces cerevisiae)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Rpd3s histone deacetylase in complex with di-nucleosome (550 MDa, Complex)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Di-nucleosome template foward (107 kDa, DNA from synthetic construct)
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Di-nucleosome (400 MDa, Complex from Xenopus laevis)