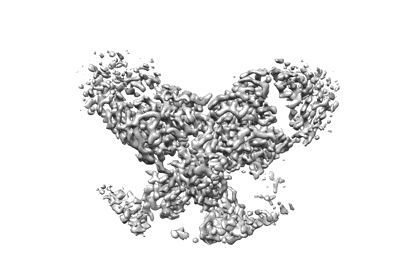
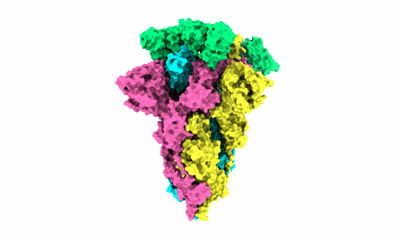
Complex of FMDV O/18074 and inter-serotype broadly neutralizing antibodies pOA-2
Resolution: 2.44 Å
EM Method: Single-particle
Fitted PDBs: 8y0q
Q-score: 0.621
Li F, Wu S, Lv L, Huang S, Zhang Z, Zerang Z, Li P, Cao Y, Bao H, Sun P, Bai X, He Y, Fu Y, Yuan H, Ma X, Zhao Z, Zhang J, Wang J, Wang T, Li D, Zhang Q, He J, Liu Z, Lu Z, Lei D, Li K
PLoS Pathog (2024) 20 pp. e1012623-e1012623 [ DOI: doi:10.1371/journal.ppat.1012623 Pubmed: 39405339 ]
- Inter-serotype broadly neutralizing antibodies poa-2 (Complex from Sus scrofa)
- Complex of fmdv o/18074 and inter-serotype broadly neutralizing antibodies poa-2 (Complex)
- Vp3 of capsid protein (23 kDa, Protein from Foot-and-mouth disease virus O)
- Vp1 of capsid protein (23 kDa, Protein from Foot-and-mouth disease virus O)
- Vp4 of capsid protein (8 kDa, Protein from Foot-and-mouth disease virus O)
- Poa2 vl (11 kDa, Protein from Sus scrofa)
- Vp2 of capsid protein (24 kDa, Protein from Foot-and-mouth disease virus O)
- Fmdv o/18074 (Complex from Foot-and-mouth disease virus O)
- Poa2 vh (13 kDa, Protein from Sus scrofa)
Complex of FMDV A/WH/CHA/09 and inter-serotype broadly neutralizing antibodies pOA-2
Resolution: 2.52 Å
EM Method: Single-particle
Fitted PDBs: 8y0r
Q-score: 0.578
Li F, Wu S, Lv L, Huang S, Zhang Z, Zerang Z, Li P, Cao Y, Bao H, Sun P, Bai X, He Y, Fu Y, Yuan H, Ma X, Zhao Z, Zhang J, Wang J, Wang T, Li D, Zhang Q, He J, Liu Z, Lu Z, Lei D, Li K
PLoS Pathog (2024) 20 pp. e1012623-e1012623 [ DOI: doi:10.1371/journal.ppat.1012623 Pubmed: 39405339 ]
- Inter-serotype broadly neutralizing antibodies poa-2 (Complex from Sus scrofa)
- Complex of fmdv a/wh/cha/09 and inter-serotype broadly neutralizing antibodies poa-2 (Complex)
- Vp2 of capsid protein (24 kDa, Protein from Foot-and-mouth disease virus A)
- Vp3 of capsid protein (24 kDa, Protein from Foot-and-mouth disease virus A)
- Fmdv a/wh/cha/09 (Complex from Foot-and-mouth disease virus A)
- Vp4 of capsid protein (8 kDa, Protein from Foot-and-mouth disease virus A)
- Vp1 of capsid protein (23 kDa, Protein from Foot-and-mouth disease virus A)
- Poa2 vl (11 kDa, Protein from Sus scrofa)
- Poa2 vh (13 kDa, Protein from Sus scrofa)
Local CryoEM structure of the SARS-CoV-2 S6P(B.1.1.529) in complex with BD55-4637 Fab
Resolution: 4.08 Å
EM Method: Single-particle
Fitted PDBs: 7wrj
Q-score: 0.322
Cao Y, Jian F, Zhang Z, Yisimayi A, Hao X, Bao L, Yuan F, Yu Y, Du S, Wang J, Xiao T, Song W, Zhang Y, Liu P, An R, Wang P, Wang Y, Yang S, Niu X, Zhang Y, Gu Q, Shao F, Hu Y, Yin W, Zheng A, Wang Y, Qin C, Jin R, Xiao J, Xie XS
Cell Rep (2022) 41 pp. 111845-111845 [ Pubmed: 36493787 DOI: doi:10.1016/j.celrep.2022.111845 ]
- The sars-cov-2 s6p(b.1.1.529) in complex with bd55-4637 fab (Complex from Homo sapiens)
- Bd55-4637h (27 kDa, Protein from Homo sapiens)
- Spike protein s1 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Bd55-4637l (24 kDa, Protein from Homo sapiens)
Local structure of BD55-3546 Fab and SARS-COV2 Delta RBD complex
Resolution: 3.28 Å
EM Method: Single-particle
Fitted PDBs: 7wry
Q-score: 0.538
Cao Y, Jian F, Zhang Z, Yisimayi A, Hao X, Bao L, Yuan F, Yu Y, Du S, Wang J, Xiao T, Song W, Zhang Y, Liu P, An R, Wang P, Wang Y, Yang S, Niu X, Zhang Y, Gu Q, Shao F, Hu Y, Yin W, Zheng A, Wang Y, Qin C, Jin R, Xiao J, Xie XS
Cell Rep (2022) 41 pp. 111845-111845 [ Pubmed: 36493787 DOI: doi:10.1016/j.celrep.2022.111845 ]
- Spike protein s1 (21 kDa, Protein from SARS coronavirus B012)
- Local structure of bd55-3546 fab and sars-cov2 delta rbd complex (Complex from Homo sapiens)
- Bd55-3546l (11 kDa, Protein from SARS coronavirus B012)
- Bd55-3546h (13 kDa, Protein from SARS coronavirus B012)
Local structure of BD55-5514 and BD55-5840 Fab and Omicron BA.1 RBD complex
Resolution: 3.42 Å
EM Method: Single-particle
Fitted PDBs: 7y0w
Q-score: 0.512
Cao Y, Jian F, Zhang Z, Yisimayi A, Hao X, Bao L, Yuan F, Yu Y, Du S, Wang J, Xiao T, Song W, Zhang Y, Liu P, An R, Wang P, Wang Y, Yang S, Niu X, Zhang Y, Gu Q, Shao F, Hu Y, Yin W, Zheng A, Wang Y, Qin C, Jin R, Xiao J, Xie XS
Cell Rep (2022) 41 pp. 111845-111845 [ Pubmed: 36493787 DOI: doi:10.1016/j.celrep.2022.111845 ]
- Bd55-5514l (11 kDa, Protein from Homo sapiens)
- Spike protein s1 (21 kDa, Protein from Homo sapiens)
- Bd55-5514h (13 kDa, Protein from Homo sapiens)
- Bd55-5840l (11 kDa, Protein from Homo sapiens)
- Local structure of bd55-5514 and bd55-5840 fab and omicron ba.1 rbd complex (Complex from Homo sapiens)
- Bd55-5840h (13 kDa, Protein from Homo sapiens)
SARS-CoV-2 BA.2 variant spike protein in complex with Fab BD55-5840
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7x6a
Q-score: 0.356
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ Pubmed: 35714668 DOI: doi:10.1038/s41586-022-04980-y ]
- Heavy chain of fab bd55-5840 (13 kDa, Protein from Homo sapiens)
- Sars-cov-2 ba.2 variant spike protein in complex with fab bd55-5840 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Fab bd55-5840 (Complex)
- Light chain of fab bd55-5840 (11 kDa, Protein from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 ba.2 variant spike protein (Complex from Homo sapiens)
SARS-CoV-2 Omicron BA.2 variant spike (state 1)
Resolution: 3.62 Å
EM Method: Single-particle
Fitted PDBs: 7xiw
Q-score: 0.316
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ Pubmed: 35714668 DOI: doi:10.1038/s41586-022-04980-y ]
SARS-CoV-2 Omicron BA.2 variant spike (state 2)
Resolution: 3.25 Å
EM Method: Single-particle
Fitted PDBs: 7xix
Q-score: 0.505
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ Pubmed: 35714668 DOI: doi:10.1038/s41586-022-04980-y ]
SARS-CoV-2 Omicron BA.3 variant spike
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 7xiy
Q-score: 0.359
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ Pubmed: 35714668 DOI: doi:10.1038/s41586-022-04980-y ]
SARS-CoV-2 Omicron BA.3 variant spike (local)
Resolution: 3.74 Å
EM Method: Single-particle
Fitted PDBs: 7xiz
Q-score: 0.392
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ Pubmed: 35714668 DOI: doi:10.1038/s41586-022-04980-y ]