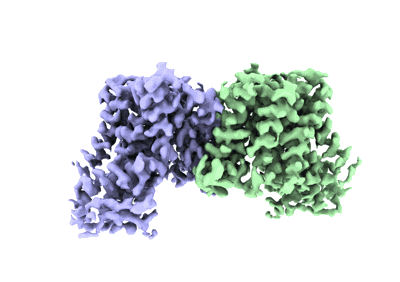
Structure of AtHKT1;1 in NaCl at 2.7 Angstroms resolution
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8w9n
Q-score: 0.633
Wang J, Luo Y, Ye F, Ding ZJ, Zheng SJ, Qiao S, Wang Y, Guo J, Yang W, Su N
Mol Plant (2024) 17 pp. 409-422 [ DOI: doi:10.1016/j.molp.2024.01.007 Pubmed: 38335958 ]
- Sodium ion (22 Da, Ligand)
- Structure of athkt1;1 in nacl at 2.7 angstroms resolution (Complex from Arabidopsis thaliana)
- Sodium transporter hkt1 (57 kDa, Protein from Arabidopsis thaliana)
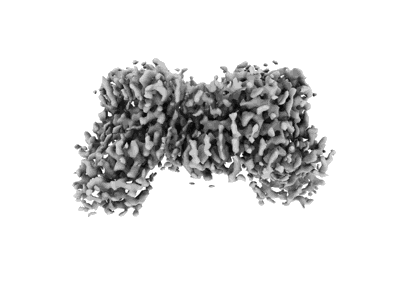
structure of AtHKT1;1 in KCl at 2.8 Angstroms resolution
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8w9o
Q-score: 0.589
Wang J, Luo Y, Ye F, Ding ZJ, Zheng SJ, Qiao S, Wang Y, Guo J, Yang W, Su N
Mol Plant (2024) 17 pp. 409-422 [ DOI: doi:10.1016/j.molp.2024.01.007 Pubmed: 38335958 ]
- Structure of athkt1;1 in kcl at 2.8 angstroms resolution (Complex from Arabidopsis thaliana)
- Potassium ion (39 Da, Ligand)
- Sodium transporter hkt1 (57 kDa, Protein from Arabidopsis thaliana)
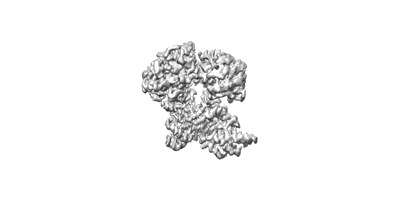
Structure of TaHKT2;1 in NaCl at 2.6 Angstroms resolution
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8w9t
Q-score: 0.617
Wang J, Luo Y, Ye F, Ding ZJ, Zheng SJ, Qiao S, Wang Y, Guo J, Yang W, Su N
Mol Plant (2024) 17 pp. 409-422 [ DOI: doi:10.1016/j.molp.2024.01.007 Pubmed: 38335958 ]
- Hkt2 (60 kDa, Protein from Triticum aestivum)
- Structure of tahkt2;1 in nacl at 2.6 angstroms resolution (Complex from Arabidopsis thaliana)
- Water (18 Da, Ligand)
- Sodium ion (22 Da, Ligand)
structure of TaHKT2;1 in KCl at 2.9 Angstroms resolution
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8w9v
Q-score: 0.598
Wang J, Luo Y, Ye F, Ding ZJ, Zheng SJ, Qiao S, Wang Y, Guo J, Yang W, Su N
Mol Plant (2024) 17 pp. 409-422 [ DOI: doi:10.1016/j.molp.2024.01.007 Pubmed: 38335958 ]
- Hkt2 (60 kDa, Protein from Triticum aestivum)
- Structure of tahkt2;1 in kcl at 2.9 angstroms resolution (Complex from Arabidopsis thaliana)
- Potassium ion (39 Da, Ligand)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7wks
Q-score: 0.527
Resolution: 2.62 Å
EM Method: Single-particle
Fitted PDBs: 7wkw
Q-score: 0.448
Resolution: 2.93 Å
EM Method: Single-particle
Fitted PDBs: 7xxb
Q-score: 0.526
Su N, Zhu A, Tao X, Ding ZJ, Chang S, Ye F, Zhang Y, Zhao C, Chen Q, Wang J, Zhou CY, Guo Y, Jiao S, Zhang S, Wen H, Ma L, Ye S, Zheng SJ, Yang F, Wu S, Guo J
Nature (2022) 609 pp. 616-621 [ DOI: doi:10.1038/s41586-022-05142-w Pubmed: 35917926 ]
- Auxin efflux carrier component 3 (73 kDa, Protein from Arabidopsis thaliana)
- Iaa-bound state atpin3(atpin3-iaa) (Complex from Arabidopsis thaliana)
- 1h-indol-3-ylacetic acid (175 Da, Ligand)
Cryo-EM structure of EDS1 and SAG101 with ATP-APDR
Resolution: 2.71 Å
EM Method: Single-particle
Fitted PDBs: 7xjp
Q-score: 0.488
Jia A, Huang S, Song W, Wang J, Meng Y, Sun Y, Xu L, Laessle H, Jirschitzka J, Hou J, Zhang T, Yu W, Hessler G, Li E, Ma S, Yu D, Gebauer J, Baumann U, Liu X, Han Z, Chang J, Parker JE, Chai J
Science (2022) 377 pp. eabq8180-eabq8180 [ DOI: doi:10.1126/science.abq8180 Pubmed: 35857644 ]
- Isopropyl alcohol (60 Da, Ligand)
- Protein eds1 (71 kDa, Protein from Arabidopsis)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Adenosine-5-diphosphoribose (559 Da, Ligand)
- Binary complex of eds1 and sag101 with molecule atp-adpr (Complex from Arabidopsis)
- Senescence-associated carboxylesterase 101 (62 kDa, Protein from Arabidopsis)
Double nuclear outer ring from the isolated yeast NPC
Resolution: 11.6 Å
EM Method: Single-particle
Fitted PDBs: 7n84
Q-score: 0.063
Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, Salzberg D, Song K, Xu C, Gumbart JC, Suslov S, Unruh J, Jaspersen SL, Chait BT, Sali A, Fernandez-Martinez J, Ludtke SJ, Villa E, Rout MP
Cell (2022) 185 pp. 361-378.e25 [ Pubmed: 34982960 DOI: doi:10.1016/j.cell.2021.12.015 ]
- Nucleoporin seh1 (39 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin 145c (81 kDa, Protein from Saccharomyces cerevisiae)
- Yeast double outer ring (10 MDa, Complex from Saccharomyces cerevisiae)
- Nucleoporin nup120 (120 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup84 (83 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup188 (188 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup133 (133 kDa, Protein from Saccharomyces cerevisiae)
- Unknown (5 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup85 (84 kDa, Protein from Saccharomyces cerevisiae)
- Protein transport protein sec13 (33 kDa, Protein from Saccharomyces cerevisiae)
Inner ring spoke from the isolated yeast NPC
Resolution: 7.6 Å
EM Method: Single-particle
Fitted PDBs: 7n85
Q-score: 0.113
Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, Salzberg D, Song K, Xu C, Gumbart JC, Suslov S, Unruh J, Jaspersen SL, Chait BT, Sali A, Fernandez-Martinez J, Ludtke SJ, Villa E, Rout MP
Cell (2022) 185 pp. 361-378.e25 [ Pubmed: 34982960 DOI: doi:10.1016/j.cell.2021.12.015 ]
- Nucleoporin nup192 (191 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin asm4 (58 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup157 (156 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup57 (57 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nic96 (96 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nsp1 (86 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup49/nsp49 (49 kDa, Protein from Saccharomyces cerevisiae)
- Unknown connectors (3 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup53 (52 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup170 (169 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup188 (188 kDa, Protein from Saccharomyces cerevisiae)
- Spoke from yeast inner ring (16 MDa, Complex from Saccharomyces cerevisiae)