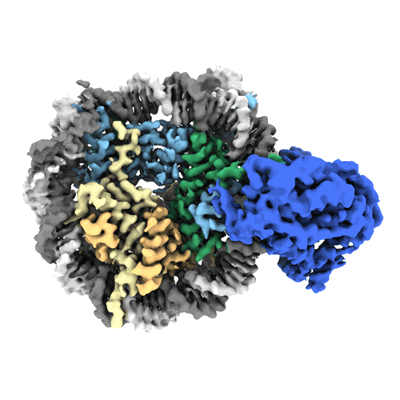
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8v4y
Q-score: 0.53
Chio US, Palovcak E, Smith AAA, Autzen H, Munoz EN, Yu Z, Wang F, Agard DA, Armache JP, Narlikar GJ, Cheng Y
Nat Commun (2024) 15 pp. 2225-2225 [ DOI: doi:10.1038/s41467-024-46178-y Pubmed: 38472177 ]
- Nucleosome with xenopus laevis histones (200 kDa, Complex)
- Swi/snf-related matrix-associated actin-dependent regulator of chromatin subfamily a member 5 (122 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Atp-dependent chromatin remodeler snf2h in complex with nucleosome (300 kDa, Complex)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Widom 601 dna (147-mer) with 60 base pairs flanking dna (forward strand) (63 kDa, DNA from synthetic construct)
- Beryllium trifluoride ion (66 Da, Ligand)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Histone h2a type 1 (13 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Widom 601 dna (147-mer) with 60 base pairs flanking dna (reverse strand) (64 kDa, DNA from synthetic construct)
- Snf2h (100 kDa, Complex from Homo sapiens)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
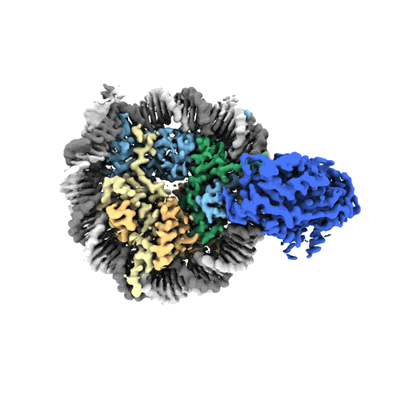
Cryo-EM structure of SNF2h-nucleosome complex (consensus structure)
Chio US, Palovcak E, Smith AAA, Autzen H, Munoz EN, Yu Z, Wang F, Agard DA, Armache JP, Narlikar GJ, Cheng Y
Nat Commun (2024) 15 pp. 2225-2225 [ DOI: doi:10.1038/s41467-024-46178-y Pubmed: 38472177 ]
- Histone h2a type 1 (Protein from Xenopus laevis)
- Nucleosome with xenopus laevis histones (Complex from Xenopus laevis)
- Snf2h (Complex from Homo sapiens)
- Histone h2b 1.1 (Protein from Xenopus laevis)
- Histone h4 (Protein from Xenopus laevis)
- Widom 601 dna (147-mer) plus 60 base pairs flanking dna reverse (DNA from synthetic construct)
- Histone h3.2 (Protein from Xenopus laevis)
- Swi/snf-related matrix-associated actin-dependent regulator of chromatin subfamily a member 5 (Protein from Homo sapiens)
- Widom 601 dna (147-mer) plus 60 base pairs flanking dna forward (DNA from synthetic construct)
- Atp-dependent chromatin remodeler snf2h in complex with nucleosome (100 kDa, Complex from Homo sapiens)
Cryo-EM structure of SNF2h-nucleosome complex (single-bound structure)
Chio US, Palovcak E, Smith AAA, Autzen H, Munoz EN, Yu Z, Wang F, Agard DA, Armache JP, Narlikar GJ, Cheng Y
Nat Commun (2024) 15 pp. 2225-2225 [ DOI: doi:10.1038/s41467-024-46178-y Pubmed: 38472177 ]
- Nucleosome with xenopus laevis histones (200 kDa, Complex)
- Histone h2a type 1 (Protein from Xenopus laevis)
- Atp-dependent chromatin remodeler snf2h in complex with nucleosome (300 kDa, Complex)
- Widom 601 dna (147-mer) plus 60 base pairs flanking dna forward strand (DNA from synthetic construct)
- Widom 601 dna (147-mer) plus 60 base pairs flanking dna reverse strand (DNA from synthetic construct)
- Histone h2b 1.1 (Protein from Xenopus laevis)
- Histone h4 (Protein from Xenopus laevis)
- Swi/snf-related matrix-associated actin-dependent regulator of chromatin subfamily a member 5 (Protein from Homo sapiens)
- Histone h3.2 (Protein from Xenopus laevis)
- Snf2h (100 kDa, Complex from Homo sapiens)
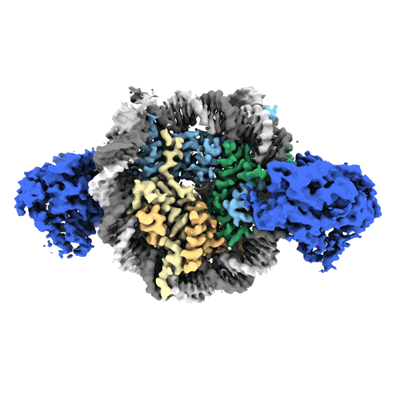
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8v6v
Q-score: 0.477
Chio US, Palovcak E, Smith AAA, Autzen H, Munoz EN, Yu Z, Wang F, Agard DA, Armache JP, Narlikar GJ, Cheng Y
Nat Commun (2024) 15 pp. 2225-2225 [ DOI: doi:10.1038/s41467-024-46178-y Pubmed: 38472177 ]
- Nucleosome with xenopus laevis histones (200 kDa, Complex)
- Swi/snf-related matrix-associated actin-dependent regulator of chromatin subfamily a member 5 (122 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Atp-dependent chromatin remodeler snf2h in complex with nucleosome (300 kDa, Complex)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Widom 601 dna (147-mer) with 60 base pairs flanking dna (forward strand) (63 kDa, DNA from synthetic construct)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Histone h2a type 1 (13 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Widom 601 dna (147-mer) with 60 base pairs flanking dna (reverse strand) (64 kDa, DNA from synthetic construct)
- Snf2h (100 kDa, Complex from Homo sapiens)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
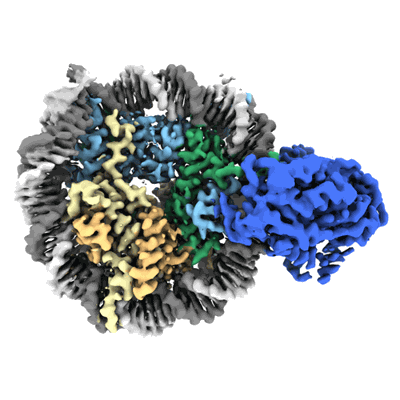
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8v7l
Q-score: 0.456
Chio US, Palovcak E, Smith AAA, Autzen H, Munoz EN, Yu Z, Wang F, Agard DA, Armache JP, Narlikar GJ, Cheng Y
Nat Commun (2024) 15 pp. 2225-2225 [ DOI: doi:10.1038/s41467-024-46178-y Pubmed: 38472177 ]
- Nucleosome with xenopus laevis histones (200 kDa, Complex)
- Swi/snf-related matrix-associated actin-dependent regulator of chromatin subfamily a member 5 (122 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Atp-dependent chromatin remodeler snf2h in complex with nucleosome (300 kDa, Complex)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Widom 601 dna (147-mer) with 60 base pairs flanking dna (forward strand) (63 kDa, DNA from synthetic construct)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Widom 601 dna (147-mer) plus 60 base pairs flanking dna (reverse strand) (64 kDa, DNA from synthetic construct)
- Histone h2a type 1 (13 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Snf2h (100 kDa, Complex from Homo sapiens)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
EM map of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8d4x
Q-score: 0.358
Wang F, He Q, Zhan W, Yu Z, Finkin-Groner E, Ma X, Lin G, Li H
Structure (2023) 31 pp. 541-552.e4 [ DOI: doi:10.1016/j.str.2023.03.010 Pubmed: 37040767 ]
- Homodimer of human e3 ligase ubr5 (610 kDa, Complex from Homo sapiens)
- E3 ubiquitin-protein ligase ubr5 (310 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
EM map of the human UBR5 HECT-type E3 ubiquitin ligase in a C2 symmetric dimeric form
Resolution: 2.66 Å
EM Method: Single-particle
Fitted PDBs: 8e0q
Q-score: 0.425
Wang F, He Q, Zhan W, Yu Z, Finkin-Groner E, Ma X, Lin G, Li H
Structure (2023) 31 pp. 541-552.e4 [ DOI: doi:10.1016/j.str.2023.03.010 Pubmed: 37040767 ]
- E3 ubiquitin-protein ligase ubr5 (310 kDa, Protein from Homo sapiens)
- Homodimer of human e3 ligase ubr5 (610 kDa, Complex from Homo sapiens)
- Zinc ion (65 Da, Ligand)
Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like environment, Saposin A nanodisc
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8equ
Q-score: 0.422
Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE
eLife (2023) 12 [ Pubmed: 36695574 DOI: doi:10.7554/eLife.84477 ]
- Saposin a, polyalanine model (6 kDa, Protein from Homo sapiens)
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (744 Da, Ligand)
- Saposin-a (11 kDa, Protein from Homo sapiens)
- Structure of sars-cov-2 orf3a in late endosome/lysosome-like environment, saposin a nanodisc (Complex from Severe acute respiratory syndrome coronavirus 2)
- Orf3a protein (36 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Shaban NM, Yan R, Shi K, Moraes SN, Cheng AZ, Carpenter MA, McLellan JS, Yu Z, Harris RS
Sci Adv (2022) 8 pp. eabm2827-eabm2827 [ DOI: doi:10.1126/sciadv.abm2827 Pubmed: 35476445 ]