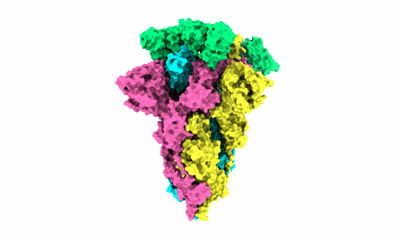
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
Resolution: 3.64 Å
EM Method: Single-particle
Fitted PDBs: 7yoy
Q-score: 0.464
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ Pubmed: 37992686 DOI: doi:10.1016/j.xcrm.2023.101296 ]
- 5e3,3e8 (Complex from Oryctolagus cuniculus)
- 5e3 heavy chain (13 kDa, Protein from Oryctolagus cuniculus)
- Ebv ghgl-gp42 in complex with mabs 3e8 and 5e3 (Complex)
- Soluble gp42 (15 kDa, Protein from Human gammaherpesvirus 4)
- Gp42 (Complex from Human gammaherpesvirus 4)
- 3e8 heavy chain (12 kDa, Protein from Oryctolagus cuniculus)
- 5e3 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- 3e8 light chain (11 kDa, Protein from Oryctolagus cuniculus)
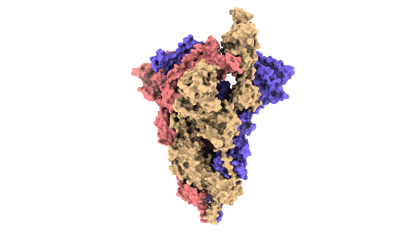
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
Resolution: 3.54 Å
EM Method: Single-particle
Fitted PDBs: 7yp1
Q-score: 0.523
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ Pubmed: 37992686 DOI: doi:10.1016/j.xcrm.2023.101296 ]
- Gh,gl (Complex from Human gammaherpesvirus 4)
- 10e4 (Complex from Oryctolagus cuniculus)
- Ebv gl (8 kDa, Protein from Human gammaherpesvirus 4)
- 10e4 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- Ebv gh (22 kDa, Protein from Human gammaherpesvirus 4)
- 10e4 heavy chain (12 kDa, Protein from Oryctolagus cuniculus)
- Ghgl-gp42 in complex with mab 10e4 (Complex)
Cryo-EM density map of EBV gHgL-gp42 in complex with four mAbs 5E3, 3E8, 6H2 and 10E4
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ Pubmed: 37992686 DOI: doi:10.1016/j.xcrm.2023.101296 ]
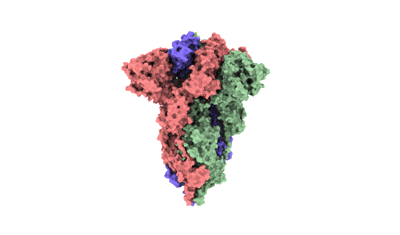
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)
Resolution: 3.52 Å
EM Method: Single-particle
Fitted PDBs: 7yp2
Q-score: 0.53
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ Pubmed: 37992686 DOI: doi:10.1016/j.xcrm.2023.101296 ]
- Gh (Complex from Human gammaherpesvirus 4)
- 6h2 (Complex from Mus musculus)
- Envelope glycoprotein h (34 kDa, Protein from Human gammaherpesvirus 4)
- 6h2 light chain (12 kDa, Protein from Mus musculus)
- Ebv ghgl-gp42 in complex with mab 6h2 (Complex)
- 6h2 heavy chain (13 kDa, Protein from Mus musculus)
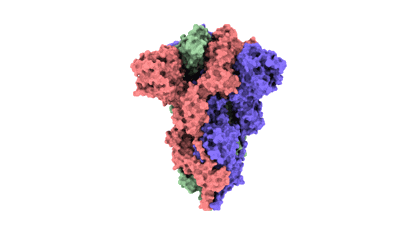
SARS-CoV-2 BA.2 variant spike protein in complex with Fab BD55-5840
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7x6a
Q-score: 0.356
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
- Fab bd55-5840 (Complex)
- Heavy chain of fab bd55-5840 (13 kDa, Protein from Homo sapiens)
- Light chain of fab bd55-5840 (11 kDa, Protein from Homo sapiens)
- Sars-cov-2 ba.2 variant spike protein in complex with fab bd55-5840 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 ba.2 variant spike protein (Complex from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 Omicron BA.2 variant spike (state 1)
Resolution: 3.62 Å
EM Method: Single-particle
Fitted PDBs: 7xiw
Q-score: 0.316
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
SARS-CoV-2 Omicron BA.2 variant spike (state 2)
Resolution: 3.25 Å
EM Method: Single-particle
Fitted PDBs: 7xix
Q-score: 0.505
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
SARS-CoV-2 Omicron BA.3 variant spike
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 7xiy
Q-score: 0.359
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
SARS-CoV-2 Omicron BA.3 variant spike (local)
Resolution: 3.74 Å
EM Method: Single-particle
Fitted PDBs: 7xiz
Q-score: 0.392
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
SARS-CoV-2 Omicron BA.4 variant spike
Resolution: 3.52 Å
EM Method: Single-particle
Fitted PDBs: 7xnq
Q-score: 0.369
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]