Cryo-EM structure of Hepatitis B virus surface antigen subviral particle with D2 symmetry
Resolution: 6.6 Å
EM Method: Single-particle
Fitted PDBs: 8ymj
Q-score: 0.233
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 8ymk
Q-score: 0.242
Cryo-EM structure of Hepatitis B virus surface antigen subviral particle with D4 symmetry
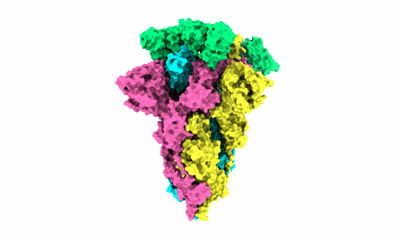
SARS-CoV-2 BA.2 variant spike protein in complex with Fab BD55-5840
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7x6a
Q-score: 0.356
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
- Fab bd55-5840 (Complex)
- Heavy chain of fab bd55-5840 (13 kDa, Protein from Homo sapiens)
- Light chain of fab bd55-5840 (11 kDa, Protein from Homo sapiens)
- Sars-cov-2 ba.2 variant spike protein in complex with fab bd55-5840 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 ba.2 variant spike protein (Complex from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
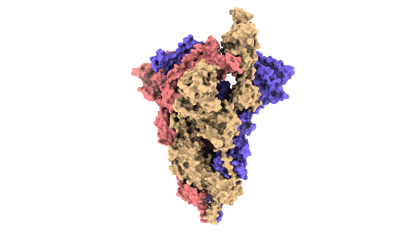
SARS-CoV-2 Omicron BA.2 variant spike (state 1)
Resolution: 3.62 Å
EM Method: Single-particle
Fitted PDBs: 7xiw
Q-score: 0.316
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
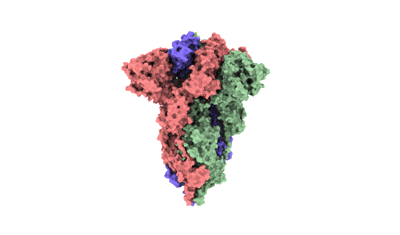
SARS-CoV-2 Omicron BA.2 variant spike (state 2)
Resolution: 3.25 Å
EM Method: Single-particle
Fitted PDBs: 7xix
Q-score: 0.505
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
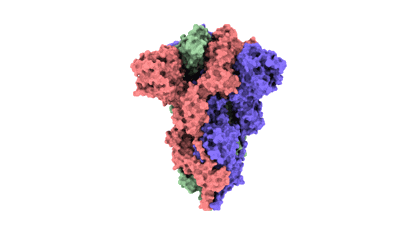
SARS-CoV-2 Omicron BA.3 variant spike
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 7xiy
Q-score: 0.359
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
SARS-CoV-2 Omicron BA.3 variant spike (local)
Resolution: 3.74 Å
EM Method: Single-particle
Fitted PDBs: 7xiz
Q-score: 0.392
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]