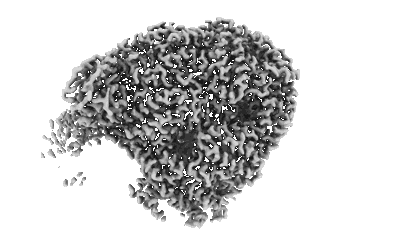
Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: B.1 variant 3 closed RBDs
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 9crc
Q-score: 0.486
Ke Z, Peacock TP, Brown JC, Sheppard CM, Croll TI, Kotecha A, Goldhill DH, Barclay WS, Briggs JAG
EMBO J (2024) 43 pp. 6469-6495 [ Pubmed: 39543395 DOI: doi:10.1038/s44318-024-00303-1 ]
Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Mu (B.1.621) variant 3 closed RBDs
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 9cri
Q-score: 0.506
Ke Z, Peacock TP, Brown JC, Sheppard CM, Croll TI, Kotecha A, Goldhill DH, Barclay WS, Briggs JAG
EMBO J (2024) 43 pp. 6469-6495 [ Pubmed: 39543395 DOI: doi:10.1038/s44318-024-00303-1 ]
ApoF at 2.2A resolution imaged at 0.55A/pixel on Falcon C
Karia D, Koh FA, Yang W, Cushing V, Basanta B, Mihaylov D, Khavnekar S, Vyroubal O, Malinsky M, Shanel O, Dolezal V, Plitzko JM, Yu L, Lander GC, Aricescu R, Greber B, Kotecha A
To Be Published
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Resolution: 2.0 Å
EM Method: Single-particle
Fitted PDBs: 8to2
Q-score: 0.592
Sauer PV, Cupellini L, Sutter M, Bondanza M, Dominguez Martin MA, Kirst H, Bina D, Koh AF, Kotecha A, Greber BJ, Nogales E, Polivka T, Mennucci B, Kerfeld CA
Sci Adv (2024) 10 pp. eadk7535-eadk7535 [ Pubmed: 38578996 DOI: doi:10.1126/sciadv.adk7535 ]
- Allophycocyanin subunit beta-18 (18 kDa, Protein from Synechocystis sp. PCC 6803)
- Allophycocyanin subunit alpha-b (17 kDa, Protein from Synechocystis sp. PCC 6803)
- Allophycocyanin alpha chain (17 kDa, Protein from Synechocystis sp. PCC 6803)
- Allophycocyanin beta chain (17 kDa, Protein from Synechocystis sp. PCC 6803)
- Phycobilisome 7.8 kda linker polypeptide, allophycocyanin-associated, core (7 kDa, Protein from Synechocystis sp. PCC 6803)
- Translation initiation factor if-2 (12 kDa, Protein from Synechocystis sp. PCC 6803)
- Orange carotenoid-binding protein (34 kDa, Protein from Synechocystis sp. PCC 6803)
- Phycobilisome rod-core linker polypeptide cpcg (28 kDa, Protein from Synechocystis sp. PCC 6803)
- Phycobiliprotein apce (100 kDa, Protein from Synechocystis sp. PCC 6803)
- Beta,beta-carotene-4,4'-dione (564 Da, Ligand)
- Phycocyanobilin (588 Da, Ligand)
- Complex of phycobilisome from synechocystis pcc 6803 bound to ocp (Complex from Synechocystis sp. (strain PCC 6803 / Kazusa))
Synechocystis PCC 6803 Phycobilisome quenched by OCP, high resolution
Sauer PV, Cupellini L, Sutter M, Bondanza M, Dominguez Martin MA, Kirst H, Bina D, Koh AF, Kotecha A, Greber BJ, Nogales E, Polivka T, Mennucci B, Kerfeld CA
Sci Adv (2024) 10 pp. eadk7535-eadk7535 [ Pubmed: 38578996 DOI: doi:10.1126/sciadv.adk7535 ]
Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement)
Resolution: 2.1 Å
EM Method: Single-particle
Fitted PDBs: 8tpj
Q-score: 0.675
Sauer PV, Cupellini L, Sutter M, Bondanza M, Dominguez Martin MA, Kirst H, Bina D, Koh AF, Kotecha A, Greber BJ, Nogales E, Polivka T, Mennucci B, Kerfeld CA
Sci Adv (2024) 10 pp. eadk7535-eadk7535 [ Pubmed: 38578996 DOI: doi:10.1126/sciadv.adk7535 ]
- Allophycocyanin alpha chain (17 kDa, Protein from Synechocystis sp. PCC 6803)
- Allophycocyanin beta chain (17 kDa, Protein from Synechocystis sp. PCC 6803)
- Phycobilisome 7.8 kda linker polypeptide, allophycocyanin-associated, core (7 kDa, Protein from Synechocystis sp. PCC 6803)
- Beta,beta-carotene-4,4'-dione (564 Da, Ligand)
- Orange carotenoid-binding protein (34 kDa, Protein from Synechocystis sp. PCC 6803)
- Phycobiliprotein apce (100 kDa, Protein from Synechocystis sp. PCC 6803)
- Phycobilisome bound to ocp (Complex from Synechocystis sp. PCC 6803)
- Phycocyanobilin (588 Da, Ligand)
- Water (18 Da, Ligand)
- Phycobilisome rod-core linker polypeptide cpcg (28 kDa, Protein from Synechocystis sp. PCC 6803)
Cryo-EM structure of CAK in complex with inhibitor ICEC0510-R
Resolution: 2.1 Å
EM Method: Single-particle
Fitted PDBs: 8p6z
Q-score: 0.687
Cushing VI, Koh AF, Feng J, Jurgaityte K, Bondke A, Kroll SHB, Barbazanges M, Scheiper B, Bahl AK, Barrett AGM, Ali S, Kotecha A, Greber BJ
Nat Commun (2024) 15 pp. 2265-2265 [ Pubmed: 38480681 DOI: doi:10.1038/s41467-024-46375-9 ]
- Cdk-activating kinase assembly factor mat1 (10 kDa, Protein from Homo sapiens)
- N7-(phenylmethyl)-3-propan-2-yl-n5-[(3r)-pyrrolidin-3-yl]pyrazolo[1,5-a]pyrimidine-5,7-diamine (350 Da, Ligand)
- Cdk-activating kinase (85 kDa, Complex from Homo sapiens)
- Cyclin-dependent kinase 7 (39 kDa, Protein from Homo sapiens)
- Cyclin-h (37 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
Cryo-EM structure of CAK in complex with inhibitor ICEC0510-S
Resolution: 2.0 Å
EM Method: Single-particle
Fitted PDBs: 8p70
Q-score: 0.68
Cushing VI, Koh AF, Feng J, Jurgaityte K, Bondke A, Kroll SHB, Barbazanges M, Scheiper B, Bahl AK, Barrett AGM, Ali S, Kotecha A, Greber BJ
Nat Commun (2024) 15 pp. 2265-2265 [ Pubmed: 38480681 DOI: doi:10.1038/s41467-024-46375-9 ]
- N7-(phenylmethyl)-3-propan-2-yl-n5-[(3s)-pyrrolidin-3-yl]pyrazolo[1,5-a]pyrimidine-5,7-diamine (350 Da, Ligand)
- Cdk-activating kinase assembly factor mat1 (10 kDa, Protein from Homo sapiens)
- Cdk-activating kinase (85 kDa, Complex from Homo sapiens)
- Cyclin-dependent kinase 7 (39 kDa, Protein from Homo sapiens)
- Cyclin-h (37 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
Cryo-EM structure of CAK in complex with inhibitor ICEC0574
Resolution: 2.0 Å
EM Method: Single-particle
Fitted PDBs: 8p71
Q-score: 0.678
Cushing VI, Koh AF, Feng J, Jurgaityte K, Bondke A, Kroll SHB, Barbazanges M, Scheiper B, Bahl AK, Barrett AGM, Ali S, Kotecha A, Greber BJ
Nat Commun (2024) 15 pp. 2265-2265 [ Pubmed: 38480681 DOI: doi:10.1038/s41467-024-46375-9 ]
- (3r,4s)-4-[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]pyrrolidin-3-ol (366 Da, Ligand)
- Cdk-activating kinase assembly factor mat1 (10 kDa, Protein from Homo sapiens)
- Cdk-activating kinase (85 kDa, Complex from Homo sapiens)
- Cyclin-dependent kinase 7 (39 kDa, Protein from Homo sapiens)
- Cyclin-h (37 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
Cryo-EM structure of CAK in complex with inhibitor ICEC0829
Resolution: 2.0 Å
EM Method: Single-particle
Fitted PDBs: 8p73
Q-score: 0.683
Cushing VI, Koh AF, Feng J, Jurgaityte K, Bondke A, Kroll SHB, Barbazanges M, Scheiper B, Bahl AK, Barrett AGM, Ali S, Kotecha A, Greber BJ
Nat Commun (2024) 15 pp. 2265-2265 [ Pubmed: 38480681 DOI: doi:10.1038/s41467-024-46375-9 ]
- (3r,4r)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidine-3,4-diol (410 Da, Ligand)
- Cdk-activating kinase assembly factor mat1 (10 kDa, Protein from Homo sapiens)
- Cdk-activating kinase (85 kDa, Complex from Homo sapiens)
- Cyclin-dependent kinase 7 (39 kDa, Protein from Homo sapiens)
- Cyclin-h (37 kDa, Protein from Homo sapiens)
- (3s,4s)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidine-3,4-diol (410 Da, Ligand)
- Water (18 Da, Ligand)