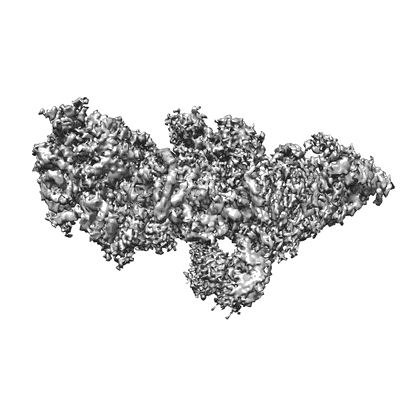
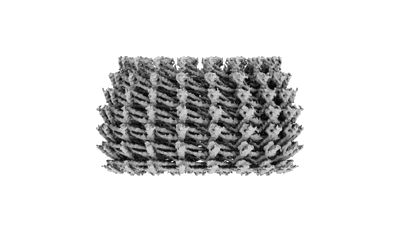
Resolution: 5.84 Å
EM Method: Single-particle
Fitted PDBs: 8dun
Q-score: 0.214
Sutton MS, Pletnev S, Callahan V, Ko S, Tsybovsky Y, Bylund T, Casner RG, Cerutti G, Gardner CL, Guirguis V, Verardi R, Zhang B, Ambrozak D, Beddall M, Lei H, Yang ES, Liu T, Henry AR, Rawi R, Schon A, Schramm CA, Shen CH, Shi W, Stephens T, Yang Y, Florez MB, Ledgerwood JE, Burke CW, Shapiro L, Fox JM, Kwong PD, Roederer M
Cell (2023) 186 pp. 2672-2689.e25 [ Pubmed: 37295404 DOI: doi:10.1016/j.cell.2023.05.019 ]
- Spike glycoprotein e1 (52 kDa, Protein from Western equine encephalitis virus)
- Spike glycoprotein e2 (45 kDa, Protein from Western equine encephalitis virus)
- Antibody skw11 heavy chain (24 kDa, Protein from Macaca fascicularis)
- Western equine encephalitis virus trimer in complex with antibody fab skw11 (Complex from Western equine encephalitis virus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Antibody skw11 light chain (23 kDa, Protein from Macaca fascicularis)
Resolution: 5.3 Å
EM Method: Helical reconstruction
Fitted PDBs: 8c5c
Q-score: 0.153
Serre L, Delaroche J, Vinit A, Schoehn G, Denarier E, Fourest-Lieuvin A, Arnal I
J Cell Sci (2023) 136 [ Pubmed: 36541084 DOI: doi:10.1242/jcs.260152 ]
- Tubulin beta chain (50 kDa, Protein from cattle)
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Taxol (853 Da, Ligand)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Tubulin alpha-1b chain (50 kDa, Protein from cattle)
- Microtubule (Complex from Bos taurus)
Mammalian Dicer in the "dicing state" with pre-miR-15a substrate
Resolution: 5.91 Å
EM Method: Single-particle
Fitted PDBs: 7zpi
Q-score: 0.106
Zapletal D, Taborska E, Pasulka J, Malik R, Kubicek K, Zanova M, Much C, Sebesta M, Buccheri V, Horvat F, Jenickova I, Prochazkova M, Prochazka J, Pinkas M, Novacek J, Joseph DF, Sedlacek R, Bernecky C, O'Carroll D, Stefl R, Svoboda P
Mol Cell (2022) 82 pp. 4064-4079.e13 [ DOI: doi:10.1016/j.molcel.2022.10.010 Pubmed: 36332606 ]
- Endoribonuclease dicer (226 kDa, Protein from Mus musculus)
- Pre-mir-15a (Complex from Mus musculus)
- Dicing state of mouse somatic dicer (Complex from Mus musculus)
- 59-nt precursor of mir-15a (19 kDa, RNA from Mus musculus)
- Dicing state of mouse somatic dicer with pre-mir-15a (Complex)
Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 1
Resolution: 5.1 Å
EM Method: Helical reconstruction
Fitted PDBs: 7ro2
Q-score: 0.255
Wang F, Cvirkaite-Krupovic V, Vos M, Beltran LC, Kreutzberger MAB, Winter JM, Su Z, Liu J, Schouten S, Krupovic M, Egelman EH
Cell (2022) 185 pp. 1297- [ Pubmed: 35325592 DOI: doi:10.1016/j.cell.2022.02.019 ]
Cryo-EM structure of the NR subunit from X. laevis NPC
Resolution: 5.6 Å
EM Method: Single-particle
Fitted PDBs: 7wb4
Q-score: 0.214
Huang G, Zhan X, Zeng C, Zhu X, Liang K, Zhao Y, Wang P, Wang Q, Zhou Q, Tao Q, Liu M, Lei J, Yan C, Shi Y
Cell Res (2022) 32 pp. 349-358 [ DOI: doi:10.1038/s41422-021-00610-w Pubmed: 35177819 ]
- Nuclear pore complex protein (105 kDa, Protein from Xenopus laevis)
- Nuclear pore complex protein nup96 (105 kDa, Protein from Xenopus laevis)
- Nuclear pore complex protein nup85 (75 kDa, Protein from Xenopus laevis)
- Nup155-prov protein (154 kDa, Protein from Xenopus laevis)
- Outer nup160 (162 kDa, Protein from Xenopus laevis)
- Protein elys (268 kDa, Protein from Xenopus laevis)
- The nr subunit (Complex from Xenopus laevis)
- Nucleoporin seh1-b (39 kDa, Protein from Xenopus laevis)
- Gator complex protein sec13 (35 kDa, Protein from Xenopus laevis)
- Mgc83295 protein (227 kDa, Protein from Xenopus laevis)
- Mgc154553 protein (41 kDa, Protein from Xenopus laevis)
- Outer nup133 (127 kDa, Protein from Xenopus laevis)
- Nuclear pore complex protein nup93 (93 kDa, Protein from Xenopus laevis)
- Mgc83926 protein (36 kDa, Protein from Xenopus laevis)
Resolution: 5.5 Å
EM Method: Single-particle
Fitted PDBs: 6lk8
Q-score: 0.089
Huang G, Zhang Y, Zhu X, Zeng C, Wang Q, Zhou Q, Tao Q, Liu M, Lei J, Yan C, Shi Y
Cell Res (2020) 30 pp. 520-531 [ Pubmed: 32376910 DOI: doi:10.1038/s41422-020-0319-4 ]
- Nup214 complex coiled coil region 2, helix 1 (2 kDa, Protein from Xenopus laevis)
- Bridge domain (33 kDa, Protein from Xenopus laevis)
- Nup214 complex coiled coil region 2, helix 2 (2 kDa, Protein from Xenopus laevis)
- Cytoplasmic ring subunit of xenopus laevis npc (Complex from Xenopus laevis)
- Nup214 complex coiled coil region 1, helix 3 (6 kDa, Protein from Xenopus laevis)
- Nup214 complex coiled coil region 1, helix 2 (6 kDa, Protein from Xenopus laevis)
- Outer nup133 (127 kDa, Protein from Xenopus laevis)
- Nup358 complex, clamps (322 kDa, Protein from Xenopus laevis)
- Nuclear pore complex protein nup85 (75 kDa, Protein from Xenopus laevis)
- Gator complex protein sec13 (35 kDa, Protein from Xenopus laevis)
- Nuclear pore complex protein (105 kDa, Protein from Xenopus laevis)
- Nuclear pore complex protein nup96 (105 kDa, Protein from Xenopus laevis)
- Mgc83295 protein (227 kDa, Protein from Xenopus laevis)
- Nucleoporin seh1-a (36 kDa, Protein from Xenopus laevis)
- Nup214 complex coiled coil region 2, helix 3 (2 kDa, Protein from Xenopus laevis)
- Outer nup160 (162 kDa, Protein from Xenopus laevis)
- Mgc154553 protein (41 kDa, Protein from Xenopus laevis)
- Mgc83926 protein (36 kDa, Protein from Xenopus laevis)
- Nup214 complex coiled-coil region 1, helix 1 (5 kDa, Protein from Xenopus laevis)
Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity
Resolution: 5.0 Å
EM Method: Single-particle
Fitted PDBs: 7o3z
Q-score: 0.168
Gupta TK, Klumpe S, Gries K, Heinz S, Wietrzynski W, Ohnishi N, Niemeyer J, Spaniol B, Schaffer M, Rast A, Ostermeier M, Strauss M, Plitzko JM, Baumeister W, Rudack T, Sakamoto W, Nickelsen J, Schuller JM, Schroda M, Engel BD
Cell (2021) 184 pp. 3643- [ DOI: doi:10.1016/j.cell.2021.05.011 Pubmed: 34166613 ]
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-15
Resolution: 5.87 Å
EM Method: Single-particle
Fitted PDBs: 7l57
Q-score: 0.311
Rapp M, Guo Y, Reddem ER, Yu J, Liu L, Wang P, Cerutti G, Katsamba P, Bimela JS, Bahna FA, Mannepalli SM, Zhang B, Kwong PD, Huang Y, Ho DD, Shapiro L, Sheng Z
Cell Rep (2021) 35 pp. 108950-108950 [ DOI: doi:10.1016/j.celrep.2021.108950 Pubmed: 33794145 ]
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab 2-15 variable domain heavy chain (14 kDa, Protein from Homo sapiens)
- Fab 2-15 variable domain light chain (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cryo-em structure of the sars-cov-2 spike glycoprotein bound to fab 2-15 (Complex from Homo sapiens)
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4
Resolution: 5.07 Å
EM Method: Single-particle
Fitted PDBs: 7l58
Q-score: 0.331
Rapp M, Guo Y, Reddem ER, Yu J, Liu L, Wang P, Cerutti G, Katsamba P, Bimela JS, Bahna FA, Mannepalli SM, Zhang B, Kwong PD, Huang Y, Ho DD, Shapiro L, Sheng Z
Cell Rep (2021) 35 pp. 108950-108950 [ DOI: doi:10.1016/j.celrep.2021.108950 Pubmed: 33794145 ]
- Sars-cov-2 trimeric spike glycoprotein bound to one copy of fab h4 (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Fab h4 variable domain heavy chain (13 kDa, Protein from Homo sapiens)
- Fab h4 variable domain light chain (12 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM map of DH851.3 bound to HIV-1 CH505 Env
Resolution: 5.6 Å
EM Method: Single-particle
Fitted PDBs: 7lu9
Q-score: 0.22
Williams WB, Meyerhoff RR, Edwards RJ, Li H, Manne K, Nicely NI, Henderson R, Zhou Y, Janowska K, Mansouri K, Gobeil S, Evangelous T, Hora B, Berry M, Abuahmad AY, Sprenz J, Deyton M, Stalls V, Kopp M, Hsu AL, Borgnia MJ, Stewart-Jones GBE, Lee MS, Bronkema N, Moody MA, Wiehe K, Bradley T, Alam SM, Parks RJ, Foulger A, Oguin T, Sempowski GD, Bonsignori M, LaBranche CC, Montefiori DC, Seaman M, Santra S, Perfect J, Francica JR, Lynn GM, Aussedat B, Walkowicz WE, Laga R, Kelsoe G, Saunders KO, Fera D, Kwong PD, Seder RA, Bartesaghi A, Shaw GM, Acharya P, Haynes BF
Cell (2021) 184 pp. 2955- [ Pubmed: 34019795 DOI: doi:10.1016/j.cell.2021.04.042 ]
- Dh851.3 heavy chain (24 kDa, Protein from Macaca mulatta)
- Envelope glycoprotein gp120 (51 kDa, Protein from Human immunodeficiency virus 1)
- Dh851.3 bound to hiv-1 ch505 env (Complex from Human immunodeficiency virus 1)
- Dh851.3 light chain (22 kDa, Protein from Macaca mulatta)
- Envelope glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Dh851.3 light chain (21 kDa, Protein from Macaca mulatta)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Dh851.3 heavy chain (23 kDa, Protein from Macaca mulatta)