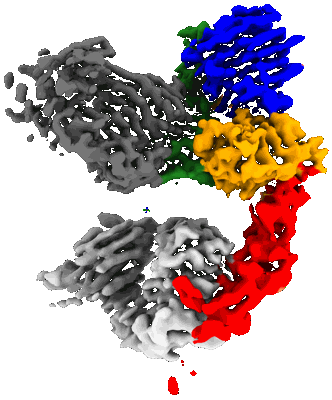
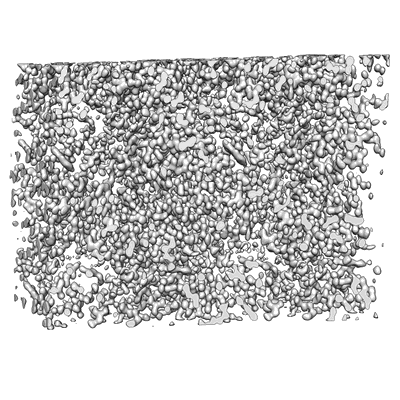
MicroED structure of bovine liver catalase with missing cone solved by suspended drop
Resolution: 4.0 Å
EM Method: Electron Crystallography
Fitted PDBs: 9bdj
Q-score: 0.483
Gillman C, Bu G, Danelius E, Hattne J, Nannenga BL, Gonen T
J Struct Biol X (2024) 9 pp. 100102-100102 [ DOI: doi:10.1016/j.yjsbx.2024.100102 Pubmed: 38962493 ]
- Nadph dihydro-nicotinamide-adenine-dinucleotide phosphate (745 Da, Ligand)
- Bovine liver catalase (Complex from Bos taurus)
- Catalase (59 kDa, Protein from Bos taurus)
- Protoporphyrin ix containing fe (616 Da, Ligand)
MicroED structure of the C11 cysteine protease clostripain
Ruma YN, Bu G, Hattne J, Gonen T
J Struct Biol X (2024) 10 pp. 100107-100107 [ DOI: doi:10.1016/j.yjsbx.2024.100107 Pubmed: 39100863 ]
- Clostripain (59 kDa, Complex from Hathewaya histolytica)
- Sodium ion (22 Da, Ligand)
- Water (18 Da, Ligand)
- Clostripain (59 kDa, Protein from Hathewaya histolytica)
Resolution: 2.15 Å
EM Method: Electron Crystallography
Fitted PDBs: 8vd7
Q-score: 0.717
Gillman C, Bu G, Danelius E, Hattne J, Nannenga BL, Gonen T
J Struct Biol X (2024) 9 pp. 100102-100102 [ DOI: doi:10.1016/j.yjsbx.2024.100102 Pubmed: 38962493 ]
- Water (18 Da, Ligand)
- Chloride ion (35 Da, Ligand)
- 3c-like proteinase nsp5 (33 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 main protease (mpro) (Complex from Severe acute respiratory syndrome coronavirus 2)
Cryogenic electron microscopy structure of human serum albumin in complex with teniposide
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8vac
Q-score: 0.463
Catalano C, Lucier KW, To D, Senko S, Tran NL, Farwell AC, Silva SM, Dip PV, Poweleit N, Scapin G
J Struct Biol (2024) 216 pp. 108105-108105 [ Pubmed: 38852682 DOI: doi:10.1016/j.jsb.2024.108105 ]
- Serum albumin (66 kDa, Protein from Homo sapiens)
- (5s,5ar,8ar,9r)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-o-(thiophen-2-ylmethylidene)-beta-d-glucopyranoside (656 Da, Ligand)
- Human serum albumin with teniposide (Complex from Homo sapiens)
Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 8vae
Q-score: 0.407
Catalano C, Lucier KW, To D, Senko S, Tran NL, Farwell AC, Silva SM, Dip PV, Poweleit N, Scapin G
J Struct Biol (2024) 216 pp. 108105-108105 [ Pubmed: 38852682 DOI: doi:10.1016/j.jsb.2024.108105 ]
- Serum albumin (66 kDa, Protein from Homo sapiens)
- Human serum albumin in complex with salicyl acid (Complex from Homo sapiens)
- 2-hydroxybenzoic acid (138 Da, Ligand)
Cryogenic electron microscopy structure of apo human serum albumin
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8vaf
Q-score: 0.464
Catalano C, Lucier KW, To D, Senko S, Tran NL, Farwell AC, Silva SM, Dip PV, Poweleit N, Scapin G
J Struct Biol (2024) 216 pp. 108105-108105 [ Pubmed: 38852682 DOI: doi:10.1016/j.jsb.2024.108105 ]
- Serum albumin (66 kDa, Protein from Homo sapiens)
- Apo human serum albumin (Complex from Homo sapiens)
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Resolution: 3.17 Å
EM Method: Single-particle
Fitted PDBs: 8pwh
Q-score: 0.417
Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic S, Mathieu M, Bressanelli S
J Struct Biol (2024) 216 pp. 108095-108095 [ DOI: doi:10.1016/j.jsb.2024.108095 Pubmed: 38723875 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ternary complex of her2-trastuzumab-pertuzumab fabs (171 kDa, Complex from Homo sapiens)
- Pertuzumab fab light chain (23 kDa, Protein from Homo sapiens)
- Trastuzumab fab heavy chain (23 kDa, Protein from Homo sapiens)
- Pertuzumab fab heavy chain (23 kDa, Protein from Homo sapiens)
- Trastuzumab fab light chain (23 kDa, Protein from Homo sapiens)
- Receptor tyrosine-protein kinase erbb-2 (68 kDa, Protein from Homo sapiens)
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8q6j
Q-score: 0.364
Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic S, Mathieu M, Bressanelli S
J Struct Biol (2024) 216 pp. 108095-108095 [ DOI: doi:10.1016/j.jsb.2024.108095 Pubmed: 38723875 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ternary complex of her2-trastuzumab-pertuzumab fabs (171 kDa, Complex from Homo sapiens)
- Pertuzumab fab light chain (23 kDa, Protein from Homo sapiens)
- Trastuzumab fab heavy chain (23 kDa, Protein from Homo sapiens)
- Pertuzumab fab heavy chain (23 kDa, Protein from Homo sapiens)
- Receptor tyrosine-protein kinase erbb-2 (68 kDa, Protein from Homo sapiens)
- Trastuzumab fab light chain (23 kDa, Protein from Homo sapiens)
MicroED structure of triclinic lysozyme
Clabbers MTB, Martynowycz MW, Hattne J, Gonen T
J Struct Biol X (2022) 6 pp. 100078-100078 [ DOI: doi:10.1016/j.yjsbx.2022.100078 Pubmed: 36507068 ]
- Lysozyme c (14 kDa, Protein from Gallus gallus)
- Nitrate ion (62 Da, Ligand)
- Water (18 Da, Ligand)
- Lysozyme (14 kDa, Complex from Gallus gallus)