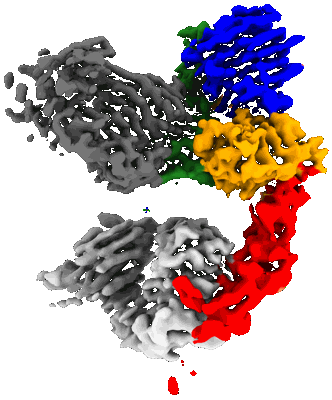
Aquaporin Z with ALFA tag and bound to nanobody
Resolution: 1.9 Å
EM Method: Single-particle
Fitted PDBs: 8uy6
Q-score: 0.68
Stover L, Bahramimoghaddam H, Wang L, Schrecke S, Yadav GP, Zhou M, Laganowsky A
J Struct Biol X (2024) 9 pp. 100097-100097 [ Pubmed: 38361954 DOI: doi:10.1016/j.yjsbx.2024.100097 ]
- Aquaporin z with alfa tag bound to nanobody (161 kDa, Complex)
- Anti-alfa nanobody (13 kDa, Protein from Vicugna pacos)
- Nanobody (Complex from Vicugna pacos)
- Aquaporin z with alfa tag (Complex from Escherichia coli)
- Cardiolipin (1 kDa, Ligand)
- Aquaporin z (26 kDa, Protein from Escherichia coli)
Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer of dimers
Oldham ML, Zuhaib Qayyum M, Kalathur RC, Rock CO, Radka CD
J Struct Biol (2024) 216 pp. 108116-108116 [ Pubmed: 39151742 DOI: doi:10.1016/j.jsb.2024.108116 ]
- Oleate hydratase (69 kDa, Protein from Staphylococcus aureus)
- Dimer of dimers of ohya (278 kDa, Complex from Staphylococcus aureus)
Oldham ML, Zuhaib Qayyum M, Kalathur RC, Rock CO, Radka CD
J Struct Biol (2024) 216 pp. 108116-108116 [ Pubmed: 39151742 DOI: doi:10.1016/j.jsb.2024.108116 ]
- Assembly of dimers of ohya bound to a liposome (977 kDa, Complex from Staphylococcus aureus)
- Oleate hydratase (69 kDa, Protein from Staphylococcus aureus)
Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex
Resolution: 2.86 Å
EM Method: Single-particle
Fitted PDBs: 8vsu
Q-score: 0.575
Chan LM, Courteau BJ, Maker A, Wu M, Basanta B, Mehmood H, Bulkley D, Joyce D, Lee BC, Mick S, Czarnik C, Gulati S, Lander GC, Verba KA
J Struct Biol (2024) 216 pp. 108108-108108 [ DOI: doi:10.1016/j.jsb.2024.108108 Pubmed: 38944401 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Serine/threonine-protein kinase stk11 (50 kDa, Protein from Homo sapiens)
- Heterotrimeric complex of serine/threonine kinase lkb1 with psuedokinase strada and scaffolding mo25a (140 kDa, Complex from Homo sapiens)
- Calcium-binding protein 39 (41 kDa, Protein from Homo sapiens)
- Isoform 3 of ste20-related kinase adapter protein alpha (47 kDa, Protein from Homo sapiens)
Apoferritin at 100 keV on Alpine detector with a side-entry cryoholder
Chan LM, Courteau BJ, Maker A, Wu M, Basanta B, Mehmood H, Bulkley D, Joyce D, Lee BC, Mick S, Czarnik C, Gulati S, Lander GC, Verba KA
J Struct Biol (2024) 216 pp. 108108-108108 [ DOI: doi:10.1016/j.jsb.2024.108108 Pubmed: 38944401 ]
- Heavy chain apoferritin from mouse (487 kDa, Complex from Mus musculus)
- Heavy chain apoferritin from mouse (Protein from Mus musculus)
Aldolase at 100 keV on the Alpine detector with a side-entry cryoholder
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Resolution: 3.17 Å
EM Method: Single-particle
Fitted PDBs: 8pwh
Q-score: 0.417
Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic S, Mathieu M, Bressanelli S
J Struct Biol (2024) 216 pp. 108095-108095 [ DOI: doi:10.1016/j.jsb.2024.108095 Pubmed: 38723875 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ternary complex of her2-trastuzumab-pertuzumab fabs (171 kDa, Complex from Homo sapiens)
- Pertuzumab fab light chain (23 kDa, Protein from Homo sapiens)
- Trastuzumab fab heavy chain (23 kDa, Protein from Homo sapiens)
- Pertuzumab fab heavy chain (23 kDa, Protein from Homo sapiens)
- Trastuzumab fab light chain (23 kDa, Protein from Homo sapiens)
- Receptor tyrosine-protein kinase erbb-2 (68 kDa, Protein from Homo sapiens)
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8q6j
Q-score: 0.364
Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic S, Mathieu M, Bressanelli S
J Struct Biol (2024) 216 pp. 108095-108095 [ DOI: doi:10.1016/j.jsb.2024.108095 Pubmed: 38723875 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ternary complex of her2-trastuzumab-pertuzumab fabs (171 kDa, Complex from Homo sapiens)
- Pertuzumab fab light chain (23 kDa, Protein from Homo sapiens)
- Trastuzumab fab heavy chain (23 kDa, Protein from Homo sapiens)
- Pertuzumab fab heavy chain (23 kDa, Protein from Homo sapiens)
- Receptor tyrosine-protein kinase erbb-2 (68 kDa, Protein from Homo sapiens)
- Trastuzumab fab light chain (23 kDa, Protein from Homo sapiens)
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8g8f
Q-score: 0.573
O'Neill AG, Burrell AL, Zech M, Elpeleg O, Harel T, Edvardson S, Mor-Shaked H, Rippert AL, Nomakuchi T, Izumi K, Kollman JM
J Biol Chem (2023) 299 pp. 105012-105012 [ Pubmed: 37414152 DOI: doi:10.1016/j.jbc.2023.105012 ]
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Inosinic acid (348 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Inosine-5'-monophosphate dehydrogenase 2 (56 kDa, Protein from Homo sapiens)
- Filament of inosine-5'-monophosphate dehydrogenase 2 - l245p mutant, bound to atp, imp, and nad+ (Complex from Homo sapiens)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8g9b
Q-score: 0.529
O'Neill AG, Burrell AL, Zech M, Elpeleg O, Harel T, Edvardson S, Mor-Shaked H, Rippert AL, Nomakuchi T, Izumi K, Kollman JM
J Biol Chem (2023) 299 pp. 105012-105012 [ Pubmed: 37414152 DOI: doi:10.1016/j.jbc.2023.105012 ]
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Inosinic acid (348 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Inosine-5'-monophosphate dehydrogenase 2 (56 kDa, Protein from Homo sapiens)
- Filament of inosine-5'-monophosphate dehydrogenase 2 - l245p mutant, bound to gtp, atp, imp, and nad+ (Complex from Homo sapiens)