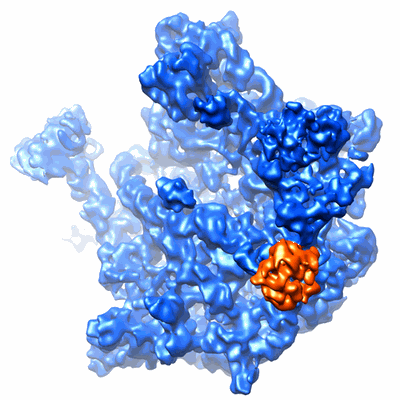
Cryo-EM structure of the 60S-Arx1-Rei1-Jjj1 complex
Greber BJ, Boehringer D, Montellese C, Ban N
Nat Struct Mol Biol (2012) 19 pp. 1228-1233 [ Pubmed: 23142985 DOI: doi:10.1038/nsmb.2425 ]
- 60s ribosomal subunit in complex with arx1, rei1, and jjj1 (2 MDa, Sample)
- Jjj1 (69 kDa, Protein from Saccharomyces cerevisiae)
- Arx1 (65 kDa, Protein from Saccharomyces cerevisiae)
- Rei1 (46 kDa, Protein from Saccharomyces cerevisiae)
- 60s ribosomal subunit (2 MDa, Complex from Saccharomyces cerevisiae)
Cryo-EM structure of the 60S-Rei1 complex
Greber BJ, Boehringer D, Montellese C, Ban N
Nat Struct Mol Biol (2012) 19 pp. 1228-1233 [ Pubmed: 23142985 DOI: doi:10.1038/nsmb.2425 ]
- Rei1 (46 kDa, Protein from Saccharomyces cerevisiae)
- 60s ribosomal subunit (2 MDa, Complex from Saccharomyces cerevisiae)
- 60s ribosomal subunit in complex with rei1 (2 MDa, Sample)
Cryo-EM structure of the 60S-Arx1-Rei1 complex
Resolution: 8.1 Å
EM Method: Single-particle
Fitted PDBs: 4v8t
Q-score: 0.135
Greber BJ, Boehringer D, Montellese C, Ban N
Nat Struct Mol Biol (2012) 19 pp. 1228-1233 [ Pubmed: 23142985 DOI: doi:10.1038/nsmb.2425 ]
- Rei1 (46 kDa, Protein from Saccharomyces cerevisiae)
- 60s ribosomal subunit (2 MDa, Complex from Saccharomyces cerevisiae)
- 60s ribosomal subunit in complex with arx1 and rei1 (2 MDa, Sample)
- Arx1 (65 kDa, Protein from Saccharomyces cerevisiae)
Structure of the E. coli methyltransferase KsgA bound to the E. coli 30S ribosomal subunit
Resolution: 13.5 Å
EM Method: Single-particle
Fitted PDBs: 4adv
Q-score: 0.061
Boehringer D, O'Farrell HC, Rife JP, Ban N
J Biol Chem (2012) 287 pp. 10453-10459 [ Pubmed: 22308031 DOI: doi:10.1074/jbc.M111.318121 ]
- E. coli methyltransferase ksga bound to the e. coli 30s ribosomal subunit (820 kDa, Sample)
- E. coli 30s ribosomal subunit (790 kDa, Complex from Escherichia coli)
- Methyltransferase ksga (30 kDa, Protein from Escherichia coli)
Structure of the E. coli 30S ribosomal subunit from a ksgA deletion strain
Boehringer D, O'Farrell HC, Rife JP, Ban N
J Biol Chem (2012) 287 pp. 10453-10459 [ DOI: doi:10.1074/jbc.M111.318121 Pubmed: 22308031 ]
- 30s ribosomal subunit (790 kDa, Complex from Escherichia coli)
- E. coli 30s ribosomal subunit from a ksga deletion strain (790 kDa, Sample)
Boehringer D, O'Farrell HC, Rife JP, Ban N
J Biol Chem (2012) 287 pp. 10453-10459 [ DOI: doi:10.1074/jbc.M111.318121 Pubmed: 22308031 ]
The Cryo-EM Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6
Resolution: 6.6 Å
EM Method: Single-particle
Fitted PDBs: 4adx
Q-score: 0.095
Greber BJ, Boehringer D, Godinic-Mikulcic V, Crnkovic A, Ibba M, Weygand-Durasevic I, Ban N
J Mol Biol (2012) 418 pp. 145-160 [ Pubmed: 22306461 DOI: doi:10.1016/j.jmb.2012.01.018 ]
Three-dimensional structure of YidC bound to the translating ribosome
Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C, Ban N
Mol Cell (2009) 34 pp. 344-353 [ DOI: doi:10.1016/j.molcel.2009.04.019 Pubmed: 19450532 ]
- Yidc (62 kDa, Protein from Escherichia coli K-12)
- Ribosome nascent chain complex (2 MDa, Complex from Escherichia coli)
- Yidc bound to a translating ribosome (2 MDa, Sample)
Three-dimensional structure of Oxa1 bound to the translating ribosome
Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C, Ban N
Mol Cell (2009) 34 pp. 344-353 [ DOI: doi:10.1016/j.molcel.2009.04.019 Pubmed: 19450532 ]
- Oxa1 (41 kDa, Protein from Saccharomyces cerevisiae)
- Ribosome nascent chain complex (2 MDa, Complex from Escherichia coli)
- Oxa1 bound to a translating ribosome (2 MDa, Sample)
Structure of hexameric DyP from Brevibacterium linens
Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N
Nat Struct Mol Biol (2008) 15 pp. 939-947 [ Pubmed: 19172747 DOI: doi:10.1038/nsmb.1473 ]
- Dye-decolorizing peroxidase (40 kDa, Protein from Brevibacterium linens)
- Structure of hexameric dyp from brevibacterium linens (240 kDa, Sample)