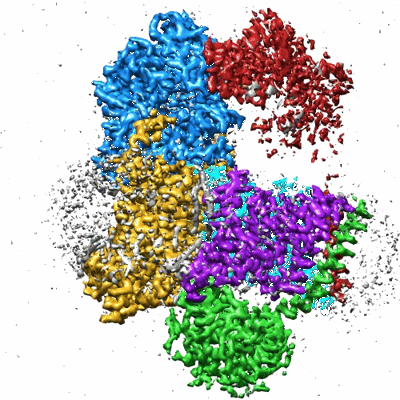
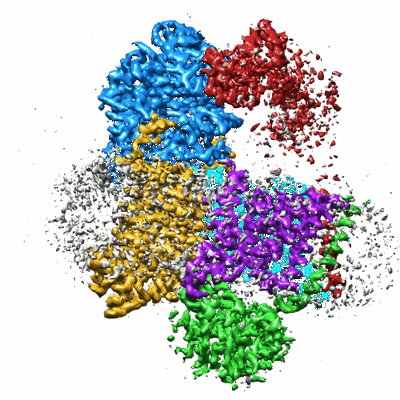
Structure of DdmD dimer with ssDNA with ADP bound
Resolution: 3.03 Å
EM Method: Single-particle
Fitted PDBs: 9khz
Q-score: 0.478
Nucleic Acids Res (2025) 53 [ DOI: doi:10.1093/nar/gkaf064 Pubmed: 39907109 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dna (5'-d(p*cp*tp*tp*tp*tp*tp*tp*tp*tp*tp*t)-3') (3 kDa, DNA from synthetic construct)
- Dna (5'-d(p*cp*cp*tp*tp*tp*tp*tp*tp*tp*tp*t)-3') (3 kDa, DNA from synthetic construct)
- Ddmd dimer with ssdna without nucleotide (300 kDa, Complex from Vibrio cholerae)
- Helicase/uvrb n-terminal domain-containing protein (136 kDa, Protein from Vibrio cholerae)
Structure of V.cholera DdmDE (2D:1E) in complex with DNA
Resolution: 3.28 Å
EM Method: Single-particle
Fitted PDBs: 9bgk
Q-score: 0.412
Yang XY, Shen Z, Wang C, Nakanishi K, Fu TM
Cell (2024) 187 pp. 5253-5266.e16 [ DOI: doi:10.1016/j.cell.2024.07.028 Pubmed: 39173632 ]
- Ddme (79 kDa, Protein from Vibrio cholerae)
- Complementary target dna (7 kDa, DNA from Vibrio cholerae)
- Helicase/uvrb n-terminal domain-containing protein (136 kDa, Protein from Vibrio cholerae)
- Guide dna (3 kDa, DNA from Vibrio cholerae)
- Non-complementary target dna (short) (3 kDa, DNA from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- Non-complementary target dna (long) (8 kDa, DNA from Vibrio cholerae)
- Ddmd dimer bind ddme in complex with dna (Complex from Vibrio cholerae)
Vibrio cholerae DdmD-DdmE holo complex
Loeff L, Adams DW, Chanez C, Stutzmann S, Righi L, Blokesch M, Jinek M
Science (2024) 385 pp. 188-194 [ DOI: doi:10.1126/science.adq0534 Pubmed: 38870273 ]
- Non-target dna strand (20 kDa, DNA from synthetic construct)
- Target dna strand (20 kDa, DNA from synthetic construct)
- Helicase/uvrb n-terminal domain-containing protein (136 kDa, Protein from Vibrio cholerae)
- Vibrio cholerae ddme (79 kDa, Protein from Vibrio cholerae)
- Ddmd-ddme holocomplex (Complex from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- 14 nucleotide dna guide with terminal 5' phosphate (4 kDa, DNA from synthetic construct)
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8u3k
Q-score: 0.545
Bravo JPK, Ramos DA, Fregoso Ocampo R, Ingram C, Taylor DW
Nature (2024) 630 pp. 961-967 [ DOI: doi:10.1038/s41586-024-07515-9 Pubmed: 38740055 ]
- Ddme (79 kDa, Protein from Vibrio cholerae)
- Dna (5'-d(p*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*t)-3') (3 kDa, DNA from synthetic construct)
- Helicase/uvrb n-terminal domain-containing protein (140 kDa, Protein from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- Ddmd apo dimer (Complex from Vibrio cholerae)
- Dna (29-mer) (8 kDa, DNA from synthetic construct)
- Dna (5'-d(p*gp*gp*ap*ap*ap*tp*gp*tp*tp*gp*ap*ap*tp*ap*c)-3') (4 kDa, DNA from synthetic construct)
DdmE in complex with guide and target DNA
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8u0j
Q-score: 0.459
Bravo JPK, Ramos DA, Fregoso Ocampo R, Ingram C, Taylor DW
Nature (2024) 630 pp. 961-967 [ DOI: doi:10.1038/s41586-024-07515-9 Pubmed: 38740055 ]
- Dna (5'-d(p*gp*tp*tp*ap*gp*ap*cp*tp*tp*tp*ap*ap*gp*t)-3') (4 kDa, DNA from synthetic construct)
- Ddme (78 kDa, Protein from Vibrio cholerae)
- Dna (5'-d(p*ap*cp*tp*tp*ap*ap*ap*gp*tp*cp*tp*ap*ap*cp*cp*tp*ap*tp*ap*gp*gp*ap*t)-3') (7 kDa, DNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Ddme (211 kDa, Complex from Streptococcus pyogenes)
- Dna (5'-d(p*ap*tp*cp*cp*tp*ap*tp*ap*gp*gp*a)-3') (3 kDa, DNA from synthetic construct)
Cryo-EM structure of Vibrio cholerae FtsE/FtsX complex
Resolution: 3.51 Å
EM Method: Single-particle
Fitted PDBs: 8tzj
Q-score: 0.471
Structure (2024) 32 pp. 188-199.e5 [ Pubmed: 38070498 DOI: doi:10.1016/j.str.2023.11.007 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Cell division atp-binding protein ftse (25 kDa, Protein from Vibrio cholerae)
- Cell division protein ftsx (36 kDa, Protein from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- Ftsex (200 kDa, Complex from Vibrio cholerae)
Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, shortened
Resolution: 3.55 Å
EM Method: Single-particle
Fitted PDBs: 8tzk
Q-score: 0.461
Structure (2024) 32 pp. 188-199.e5 [ Pubmed: 38070498 DOI: doi:10.1016/j.str.2023.11.007 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Cell division atp-binding protein ftse (25 kDa, Protein from Vibrio cholerae)
- Peptidase m23 (43 kDa, Protein from Vibrio cholerae)
- Cell division protein ftsx (36 kDa, Protein from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- Ftsex (200 kDa, Complex from Vibrio cholerae)
Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, full-length
Resolution: 3.55 Å
EM Method: Single-particle
Fitted PDBs: 8tzl
Q-score: 0.408
Structure (2024) 32 pp. 188-199.e5 [ Pubmed: 38070498 DOI: doi:10.1016/j.str.2023.11.007 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Cell division atp-binding protein ftse (25 kDa, Protein from Vibrio cholerae)
- Peptidase m23 (43 kDa, Protein from Vibrio cholerae)
- Cell division protein ftsx (36 kDa, Protein from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- Ftsex (200 kDa, Complex from Vibrio cholerae)
Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2
Resolution: 2.86 Å
EM Method: Single-particle
Fitted PDBs: 8a1u
Q-score: 0.558
Hau JL, Kaltwasser S, Muras V, Casutt MS, Vohl G, Claussen B, Steffen W, Leitner A, Bill E, Cutsail 3rd GE, DeBeer S, Vonck J, Steuber J, Fritz G
Nat Struct Mol Biol (2023) 30 pp. 1686-1694 [ Pubmed: 37710014 DOI: doi:10.1038/s41594-023-01099-0 ]
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae)
- Nqr complex with substrates nadh and q2 (220 kDa, Complex from Vibrio cholerae)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- 1,4-dihydronicotinamide adenine dinucleotide (665 Da, Ligand)
- Water (18 Da, Ligand)
- Sodium ion (22 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit a (51 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae)
- Ubiquinone-2 (318 Da, Ligand)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
Sodium pumping NADH-quinone oxidoreductase
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 8a1t
Q-score: 0.452
Hau JL, Kaltwasser S, Muras V, Casutt MS, Vohl G, Claussen B, Steffen W, Leitner A, Bill E, Cutsail 3rd GE, DeBeer S, Vonck J, Steuber J, Fritz G
Nat Struct Mol Biol (2023) 30 pp. 1686-1694 [ Pubmed: 37710014 DOI: doi:10.1038/s41594-023-01099-0 ]
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit a (48 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae)
- Riboflavin (376 Da, Ligand)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- Potassium ion (39 Da, Ligand)
- Sodium ion (22 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae)
- Nqr complex (220 kDa, Complex from Vibrio cholerae)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)