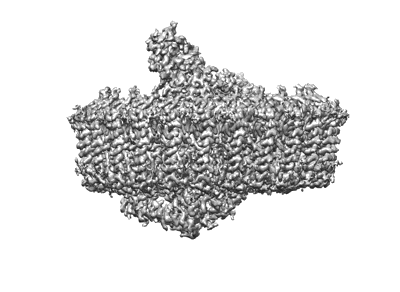
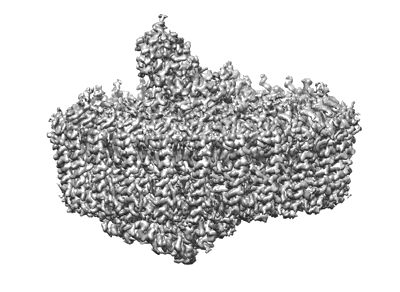
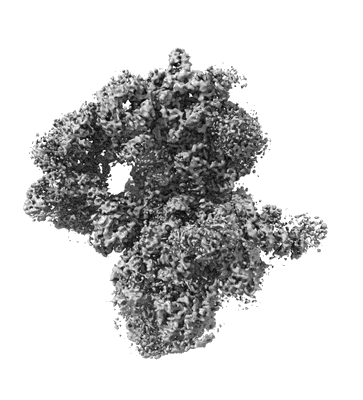
Sub-tomogram averaged structure of HIV-1 Envelope protein in native membrane
Resolution: 9.1 Å
EM Method: Subtomogram averaging
Fitted PDBs: 7ska
Q-score: 0.067
Mangala Prasad V, Leaman DP, Lovendahl KN, Croft JT, Benhaim MA, Hodge EA, Zwick MB, Lee KK
Cell (2022) 185 pp. 641-653.e17 [ Pubmed: 35123651 DOI: doi:10.1016/j.cell.2022.01.013 ]
- Human immunodeficiency virus 1 (430 kDa, Virus from Human immunodeficiency virus 1)
- Envelope glycoprotein gp120 (52 kDa, Protein from Human immunodeficiency virus 1)
- Alpha-l-fucopyranose (164 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Envelope glycoprotein gp41 (16 kDa, Protein from Human immunodeficiency virus 1)
Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A
Resolution: 2.47 Å
EM Method: Single-particle
Fitted PDBs: 7o0w
Q-score: 0.624
Qian P, Gardiner AT, Simova I, Naydenova K, Croll TI, Jackson PJ, Kloz M, Cubakova P, Kuzma M, Zeng Y, Castro-Hartmann P, van Knippenberg B, Goldie KN, Kaftan D, Hrouzek P, Hajek J, Agirre J, Siebert CA, Bina D, Sader K, Stahlberg H, Sobotka R, Russo CJ, Polivka T, Hunter CN, Koblizek M
Sci Adv (2022) 8 pp. eabk3139-eabk3139 [ DOI: doi:10.1126/sciadv.abk3139 Pubmed: 35171663 ]
- Rc-u (13 kDa, Protein from Gemmatimonas phototrophica)
- Lhc domain-containing protein (7 kDa, Protein from Gemmatimonas phototrophica)
- Multiheme_cytc domain-containing protein (38 kDa, Protein from Gemmatimonas phototrophica)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Lhh-alpha (6 kDa, Protein from Gemmatimonas phototrophica)
- Rc-hc (20 kDa, Protein from Gemmatimonas phototrophica)
- Bacteriochlorophyll a (911 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- Photosynthetic reaction center l subunit (30 kDa, Protein from Gemmatimonas phototrophica)
- (2r,5r,11r,14r)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate (1 kDa, Ligand)
- Rc-s (21 kDa, Protein from Gemmatimonas phototrophica)
- [(2~{s})-3-[(2~{r},3~{r},4~{s},5~{s},6~{r})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2-octadecanoyloxy-propyl] octadecanoate (787 Da, Ligand)
- Light-harvesting protein b:885 subunit beta (5 kDa, Protein from Gemmatimonas phototrophica)
- Rc-dlh model_1b (800 kDa, Complex from Gemmatimonas phototrophica)
- (19r,22s)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9z)-hexadec-9-enoate (689 Da, Ligand)
- Rc-m (41 kDa, Protein from Gemmatimonas phototrophica)
- Prch domain-containing protein (7 kDa, Protein from Gemmatimonas phototrophica)
- Menaquinone 8 (717 Da, Ligand)
- Bacteriopheophytin a (889 Da, Ligand)
- [(2~{s})-3-[(2~{r},3~{r},4~{r},5~{s},6~{r})-6-(hydroxymethyl)-5-[(2~{r},3~{r},4~{s},5~{s},6~{r})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]oxy-2-(12-methyltridecanoyloxy)propyl] 12-methyltridecanoate (837 Da, Ligand)
- (2~{s},3~{s},4~{s},5~{s})-4,5-diacetyloxy-3-oxidanyl-oxane-2-carboxylic acid (262 Da, Ligand)
- (2~{e},4~{e},6~{e},10~{e},12~{e},14~{e},16~{e},18~{e},20~{e},22~{z},24~{e},26~{e},28~{e})-23-methanoyl-31-methoxy-2,6,10,14,19,27,31-heptamethyl-dotriaconta-2,4,6,10,12,14,16,18,20,22,24,26,28-tridecaenoic acid (610 Da, Ligand)
- Heme c (618 Da, Ligand)
- Fe (iii) ion (55 Da, Ligand)
- Spirilloxanthin (596 Da, Ligand)
- Water (18 Da, Ligand)
- 2-acetamido-2-deoxy-alpha-d-glucopyranose (221 Da, Ligand)
- Lhc domain-containing protein (7 kDa, Protein from Gemmatimonas phototrophica)
Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A
Resolution: 2.44 Å
EM Method: Single-particle
Fitted PDBs: 7o0x
Q-score: 0.629
Qian P, Gardiner AT, Simova I, Naydenova K, Croll TI, Jackson PJ, Kloz M, Cubakova P, Kuzma M, Zeng Y, Castro-Hartmann P, van Knippenberg B, Goldie KN, Kaftan D, Hrouzek P, Hajek J, Agirre J, Siebert CA, Bina D, Sader K, Stahlberg H, Sobotka R, Russo CJ, Polivka T, Hunter CN, Koblizek M
Sci Adv (2022) 8 pp. eabk3139-eabk3139 [ DOI: doi:10.1126/sciadv.abk3139 Pubmed: 35171663 ]
- Rc-u (13 kDa, Protein from Gemmatimonas phototrophica)
- Lhc domain-containing protein (7 kDa, Protein from Gemmatimonas phototrophica)
- Multiheme_cytc domain-containing protein (38 kDa, Protein from Gemmatimonas phototrophica)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Lhh-alpha (6 kDa, Protein from Gemmatimonas phototrophica)
- Rc-hc (20 kDa, Protein from Gemmatimonas phototrophica)
- Bacteriochlorophyll a (911 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- Photosynthetic reaction center l subunit (30 kDa, Protein from Gemmatimonas phototrophica)
- (2r,5r,11r,14r)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate (1 kDa, Ligand)
- Rc-s (21 kDa, Protein from Gemmatimonas phototrophica)
- [(2~{s})-3-[(2~{r},3~{r},4~{s},5~{s},6~{r})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2-octadecanoyloxy-propyl] octadecanoate (787 Da, Ligand)
- Light-harvesting protein b:885 subunit beta (5 kDa, Protein from Gemmatimonas phototrophica)
- (19r,22s)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9z)-hexadec-9-enoate (689 Da, Ligand)
- Rc-m (41 kDa, Protein from Gemmatimonas phototrophica)
- Prch domain-containing protein (7 kDa, Protein from Gemmatimonas phototrophica)
- Menaquinone 8 (717 Da, Ligand)
- Bacteriopheophytin a (889 Da, Ligand)
- [(2~{s})-3-[(2~{r},3~{r},4~{r},5~{s},6~{r})-6-(hydroxymethyl)-5-[(2~{r},3~{r},4~{s},5~{s},6~{r})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]oxy-2-(12-methyltridecanoyloxy)propyl] 12-methyltridecanoate (837 Da, Ligand)
- (2~{s},3~{s},4~{s},5~{s})-4,5-diacetyloxy-3-oxidanyl-oxane-2-carboxylic acid (262 Da, Ligand)
- (2~{e},4~{e},6~{e},10~{e},12~{e},14~{e},16~{e},18~{e},20~{e},22~{z},24~{e},26~{e},28~{e})-23-methanoyl-31-methoxy-2,6,10,14,19,27,31-heptamethyl-dotriaconta-2,4,6,10,12,14,16,18,20,22,24,26,28-tridecaenoic acid (610 Da, Ligand)
- Heme c (618 Da, Ligand)
- Fe (iii) ion (55 Da, Ligand)
- Spirilloxanthin (596 Da, Ligand)
- Rc-dlh model_2b (800 kDa, Complex from Gemmatimonas phototrophica)
- Water (18 Da, Ligand)
- 2-acetamido-2-deoxy-alpha-d-glucopyranose (221 Da, Ligand)
- Lhc domain-containing protein (7 kDa, Protein from Gemmatimonas phototrophica)
Structure of the in situ yeast NPC
Resolution: 37.0 Å
EM Method: Subtomogram averaging
Fitted PDBs: 7n9f
Q-score: 0.043
Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, Salzberg D, Song K, Xu C, Gumbart JC, Suslov S, Unruh J, Jaspersen SL, Chait BT, Sali A, Fernandez-Martinez J, Ludtke SJ, Villa E, Rout MP
Cell (2022) 185 pp. 361-378.e25 [ Pubmed: 34982960 DOI: doi:10.1016/j.cell.2021.12.015 ]
- Nucleoporin nic96 (96 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup82 (82 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup170 (169 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup159 (159 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin seh1 (39 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nsp1 (86 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup192 (191 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup53 (52 kDa, Protein from Saccharomyces cerevisiae)
- Dynein light chain 1, cytoplasmic (10 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup133 (133 kDa, Protein from Saccharomyces cerevisiae)
- Protein transport protein sec13 (33 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin 145c (81 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup84 (83 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup188 (188 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup120 (120 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup49/nsp49 (49 kDa, Protein from Saccharomyces cerevisiae)
- Yeast npc (52 MDa, Cellular component from Saccharomyces cerevisiae)
- Nucleoporin asm4 (58 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup57 (57 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup85 (84 kDa, Protein from Saccharomyces cerevisiae)
- Orphans bound to nup192 ntd (3 kDa, Protein from Saccharomyces cerevisiae)
- Nucleoporin nup157 (156 kDa, Protein from Saccharomyces cerevisiae)
Cryo-EM structure of the Rhodospirillum rubrum RC-LH1 complex
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 7oy8
Q-score: 0.551
Qian P, Croll TI, Swainsbury DJK, Castro-Hartmann P, Moriarty NW, Sader K, Hunter CN
Biochem J (2021) 478 pp. 3253-3263 [ Pubmed: 34402504 DOI: doi:10.1042/BCJ20210511 ]
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Photosynthetic reaction center, h-chain (27 kDa, Protein from Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIMB 8255 / S1))
- Phosphate ion (94 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- (2r,5r,11r,14r)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate (1 kDa, Ligand)
- Photosynthetic reaction center l subunit (30 kDa, Protein from Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIMB 8255 / S1))
- Ubiquinone-10 (863 Da, Ligand)
- Trans-geranyl bacteriochlorophyll a (901 Da, Ligand)
- Reaction center protein m chain (34 kDa, Protein from Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIMB 8255 / S1))
- 2-azanyl-5-[(2~{e},6~{e},8~{e},10~{e},12~{e},14~{e},18~{e},22~{e},26~{e},30~{e},34~{e})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione (844 Da, Ligand)
- Bacteriopheophytin a (889 Da, Ligand)
- Light-harvesting protein b-870 beta chain (6 kDa, Protein from Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIMB 8255 / S1))
- Antenna complex, alpha/beta subunit (5 kDa, Protein from Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIMB 8255 / S1))
- Fe (iii) ion (55 Da, Ligand)
- Spirilloxanthin (596 Da, Ligand)
- Rc-lh1 from rhodospirillum rubrum (297 MDa, Complex from Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIMB 8255 / S1))
- Water (18 Da, Ligand)
- Chloride ion (35 Da, Ligand)
- Antenna complex, alpha/beta subunit (7 kDa, Protein from Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIMB 8255 / S1))
STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF THIORHODOVIBRIO STRAIN 970
Resolution: 2.82 Å
EM Method: Single-particle
Fitted PDBs: 7c9r
Q-score: 0.632
Tani K, Kanno R, Makino Y, Hall M, Takenouchi M, Imanishi M, Yu LJ, Overmann J, Madigan MT, Kimura Y, Mizoguchi A, Humbel BM, Wang-Otomo ZY
Nat Commun (2020) 11 pp. 4955-4955 [ DOI: doi:10.1038/s41467-020-18748-3 Pubmed: 33009385 ]
- Reaction center protein m chain (36 kDa, Protein from Thiorhodovibrio sp. 970)
- Light-harvesting-reaction center complex from the purple phototrophic bacterium thiorhodovibrio strain 970 (400 kDa, Complex from Thiorhodovibrio sp. 970)
- Photosynthetic reaction center, subunit h, bacterial (28 kDa, Protein from Thiorhodovibrio sp. 970)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Thiorhodovibrio sp. 970)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Alpha subunit 2 of light-harvesting 1 complex (9 kDa, Protein from Thiorhodovibrio sp. 970)
- Magnesium ion (24 Da, Ligand)
- Octadecane (254 Da, Ligand)
- (6~{e},8~{e},10~{e},12~{e},14~{e},16~{e},18~{e},20~{e},22~{e},24~{e},26~{e})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26-undecaene (600 Da, Ligand)
- Photosynthetic reaction center cytochrome c subunit (42 kDa, Protein from Thiorhodovibrio sp. 970)
- Lhc domain-containing protein (8 kDa, Protein from Thiorhodovibrio sp. 970)
- Calcium ion (40 Da, Ligand)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Thiorhodovibrio sp. 970)
- L subunit of the reaction center (30 kDa, Protein from Thiorhodovibrio sp. 970)
- Bacteriochlorophyll a (911 Da, Ligand)
- Cardiolipin (1 kDa, Ligand)
- Menaquinone 8 (717 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Bacteriopheophytin a (889 Da, Ligand)
- Ubiquinone-8 (727 Da, Ligand)
- (1r)-2-{[{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl (11e)-octadec-11-enoate (749 Da, Ligand)
- Fe (iii) ion (55 Da, Ligand)
- Diacyl glycerol (625 Da, Ligand)
- Alpha subunit 1 of light-harvesting 1 complex (8 kDa, Protein from Thiorhodovibrio sp. 970)
- Antenna complex alpha/beta subunit (9 kDa, Protein from Thiorhodovibrio sp. 970)
Virus-like Particles based on Potato Virus Y
Resolution: 4.1 Å
EM Method: Helical reconstruction
Fitted PDBs: 6hxz
Q-score: 0.447
Kezar A, Kavcic L, Polak M, Novacek J, Gutierrez-Aguirre I, Znidaric MT, Coll A, Stare K, Gruden K, Ravnikar M, Pahovnik D, Zagar E, Merzel F, Anderluh G, Podobnik M
Sci Adv (2019) 5 pp. eaaw3808-eaaw3808 [ DOI: doi:10.1126/sciadv.aaw3808 Pubmed: 31328164 ]
- Polyprotein (29 kDa, Protein from Potato virus Y)
- Coat protein of potato virus y (299 kDa, Complex from Potato virus Y)
- Virus-like particle based on potato virus y (Complex from Potato virus Y)
human Bact spliceosome core structure
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6ff4
Q-score: 0.443
Haselbach D, Komarov I, Agafonov DE, Hartmuth K, Graf B, Dybkov O, Urlaub H, Kastner B, Luhrmann R, Stark H
Cell (2018) 172 pp. 454-464.e11 [ Pubmed: 29361316 DOI: doi:10.1016/j.cell.2018.01.010 ]
- Bud13 homolog (70 kDa, Protein from Homo sapiens)
- Protein bud31 homolog (17 kDa, Protein from Homo sapiens)
- U5 snrna (36 kDa, RNA from Homo sapiens)
- Splicing factor 3b subunit 3 (135 kDa, Protein from Homo sapiens)
- Pre mrna (153 kDa, RNA from Homo sapiens)
- Cell division cycle 5-like protein (92 kDa, Protein from Homo sapiens)
- Splicing factor 3b subunit 6 (14 kDa, Protein from Homo sapiens)
- Crooked neck-like protein 1 (100 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Pre-mrna-processing factor 17 (65 kDa, Protein from Homo sapiens)
- 116 kda u5 small nuclear ribonucleoprotein component (109 kDa, Protein from Homo sapiens)
- Splicing factor 3b subunit 1 (146 kDa, Protein from Homo sapiens)
- Rna-binding motif protein, x-linked 2 (37 kDa, Protein from Homo sapiens)
- Pleiotropic regulator 1 (57 kDa, Protein from Homo sapiens)
- U6 snrna (34 kDa, RNA from Homo sapiens)
- Splicing factor 3b subunit 5 (10 kDa, Protein from Homo sapiens)
- Splicing factor 3a subunit 2 (49 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor rbm22 (46 kDa, Protein from Homo sapiens)
- Serine/arginine repetitive matrix protein 2 (300 kDa, Protein from Homo sapiens)
- Peptidyl-prolyl cis-trans isomerase-like 1 (18 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Serine/arginine repetitive matrix protein 1 (102 kDa, Protein from Homo sapiens)
- Splicing factor 3b subunit 2 (100 kDa, Protein from Homo sapiens)
- Pre-mrna-processing-splicing factor 8 (273 kDa, Protein from Homo sapiens)
- Ring finger protein 113a (38 kDa, Protein from Homo sapiens)
- Human bact spliceosome state 1 unmasked (4 MDa, Complex from Homo sapiens)
- Spliceosome-associated protein cwc15 homolog (26 kDa, Protein from Homo sapiens)
- Inositol hexakisphosphate (660 Da, Ligand)
- Phd finger-like domain-containing protein 5a (12 kDa, Protein from Homo sapiens)
- U2 snrna (60 kDa, RNA from Homo sapiens)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Peptidyl-prolyl cis-trans isomerase cwc27 homolog (53 kDa, Protein from Homo sapiens)
- Snw domain-containing protein 1 (61 kDa, Protein from Homo sapiens)
Cryo-EM structure of the T4 tail tube
Resolution: 3.4 Å
EM Method: Helical reconstruction
Fitted PDBs: 5w5f
Q-score: 0.353
Zheng W, Wang F, Taylor NMI, Guerrero-Ferreira RC, Leiman PG, Egelman EH
Structure (2017) 25 pp. 1436-1441.e2 [ Pubmed: 28757144 DOI: doi:10.1016/j.str.2017.06.017 ]
Resolution: 15.6 Å
EM Method: Single-particle
Fitted PDBs: 5tre
Q-score: 0.016
Galeano BK, Ranatunga W, Gakh O, Smith DY, Thompson JR, Isaya G
Metallomics (2017) 9 pp. 773-801 [ DOI: doi:10.1039/c7mt00089h Pubmed: 28548666 ]
- Frataxin homolog, mitochondrial (13 kDa, Protein from Saccharomyces cerevisiae)
- Iron sulfur cluster assembly protein 1, mitochondrial (15 kDa, Protein from Saccharomyces cerevisiae)
- Yfh1-isu1 (700 kDa, Complex from Saccharomyces cerevisiae)