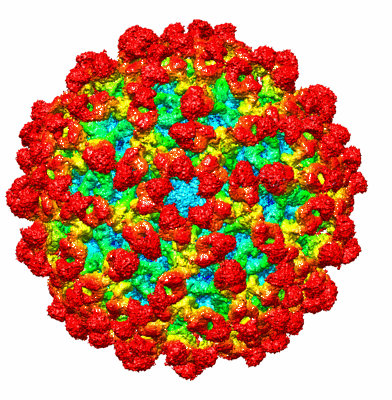
Structure of dengue virus (DENV2) in complex with prM12, an anti-PrM monoclonal antibody
Resolution: 10.2 Å
EM Method: Single-particle
Fitted PDBs: 8fe3
Q-score: 0.059
A Dowd K, Sirohi D, D Speer S, VanBlargan LA, Chen RE, Mukherjee S, Whitener BM, Govero J, Aleshnick M, Larman B, Sukupolvi-Petty S, Sevvana M, Miller AS, Klose T, Zheng A, Koenig S, Kielian M, Kuhn RJ, Diamond MS, Pierson TC
PNAS (2023) 120 pp. e2218899120-e2218899120 [ Pubmed: 36638211 DOI: doi:10.1073/pnas.2218899120 ]
- Dengue virus 2 (Virus from Dengue virus 2)
- Prm12 fab heavy chain (24 kDa, Protein from Mus musculus)
- Envelope protein e (43 kDa, Protein from Dengue virus type 2)
- Prm12 fab light chain (23 kDa, Protein from Mus musculus)
- Prm protein (9 kDa, Protein from Dengue virus type 2)
Structure of dengue virus (DENV2) in complex with prM13, an anti-PrM monoclonal antibody
Resolution: 9.8 Å
EM Method: Single-particle
Fitted PDBs: 8fe4
Q-score: 0.062
A Dowd K, Sirohi D, D Speer S, VanBlargan LA, Chen RE, Mukherjee S, Whitener BM, Govero J, Aleshnick M, Larman B, Sukupolvi-Petty S, Sevvana M, Miller AS, Klose T, Zheng A, Koenig S, Kielian M, Kuhn RJ, Diamond MS, Pierson TC
PNAS (2023) 120 pp. e2218899120-e2218899120 [ Pubmed: 36638211 DOI: doi:10.1073/pnas.2218899120 ]
- Dengue virus 2 (Virus from Dengue virus 2)
- Prm13 fab light chain (23 kDa, Protein from Mus musculus)
- Prm13 fab heavy chain (23 kDa, Protein from Mus musculus)
- Envelope protein e (43 kDa, Protein from Dengue virus type 2)
- Prm protein (9 kDa, Protein from Dengue virus type 2)
Not published ()
Localized reconstruction of EEEV
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Late pre-fusion state of EEEV (pH 5.5) with localized reconstruction
Resolution: 14.1 Å
EM Method: Single-particle
Fitted PDBs: 7n69
Q-score: 0.081
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
EEEV in complex with Fab22 at acidic pH (pH 5.5)
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
EEEV in complex with Fab22 at neutral pH (pH 7.4)
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Early pre-fusion state of EEEV (pH 5.5) with localized reconstruction
Resolution: 14.3 Å
EM Method: Single-particle
Fitted PDBs: 7n6a
Q-score: 0.069
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]