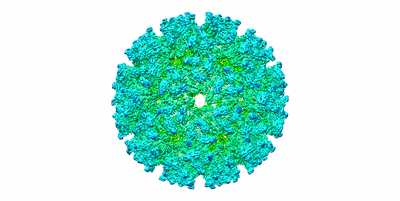
Localized reconstruction of EEEV
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Late pre-fusion state of EEEV (pH 5.5) with localized reconstruction
Resolution: 14.1 Å
EM Method: Single-particle
Fitted PDBs: 7n69
Q-score: 0.081
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
EEEV in complex with Fab22 at acidic pH (pH 5.5)
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
EEEV in complex with Fab22 at neutral pH (pH 7.4)
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Early pre-fusion state of EEEV (pH 5.5) with localized reconstruction
Resolution: 14.3 Å
EM Method: Single-particle
Fitted PDBs: 7n6a
Q-score: 0.069
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Native Mayaro virus cryo EM map, resolution 4.8 Angstroms
Resolution: 4.8 Å
EM Method: Single-particle
Fitted PDBs: 6w2u
Q-score: 0.439
Powell LA, Miller A, Fox JM, Kose N, Klose T, Kim AS, Bombardi R, Tennekoon RN, Dharshan de Silva A, Carnahan RH, Diamond MS, Rossmann MG, Kuhn RJ, Crowe Jr JE
Cell Host Microbe (2020) 28 pp. 699- [ DOI: doi:10.1016/j.chom.2020.07.008 Pubmed: 32783883 ]
- Spike glycoprotein e1 (41 kDa, Protein from Mayaro virus (strain Brazil))
- Native mayaro virus (Complex)
- Spike glycoprotein e2 (38 kDa, Protein from Mayaro virus (strain Brazil))
- Mayaro virus (Complex from Mayaro virus)
Resolution: 6.33 Å
EM Method: Single-particle
Fitted PDBs: 6vyv
Q-score: 0.339
Powell LA, Miller A, Fox JM, Kose N, Klose T, Kim AS, Bombardi R, Tennekoon RN, Dharshan de Silva A, Carnahan RH, Diamond MS, Rossmann MG, Kuhn RJ, Crowe Jr JE
Cell Host Microbe (2020) 28 pp. 699-711.e7 [ DOI: doi:10.1016/j.chom.2020.07.008 Pubmed: 32783883 ]
- Ross river virus (strain t48) (Virus from Ross river virus (strain T48))
- Fab chk-265 light chain (22 kDa, Protein from Homo sapiens)
- E2 glycoprotein (38 kDa, Protein from Ross river virus (strain T48))
- Fab chk-265 heavy chain (23 kDa, Protein from Homo sapiens)
- E1 glycoprotein (42 kDa, Protein from Ross river virus (strain T48))
Resolution: 5.3 Å
EM Method: Single-particle
Fitted PDBs: 6w09
Q-score: 0.34
Powell LA, Miller A, Fox JM, Kose N, Klose T, Kim AS, Bombardi R, Tennekoon RN, Dharshan de Silva A, Carnahan RH, Diamond MS, Rossmann MG, Kuhn RJ, Crowe Jr JE
Cell Host Microbe (2020) 28 pp. 699-711.e7 [ DOI: doi:10.1016/j.chom.2020.07.008 Pubmed: 32783883 ]
- Fab chk-265 light chain (22 kDa, Protein from Homo sapiens)
- E3 (6 kDa, Protein from Chikungunya virus)
- Chikungunya virus (Virus from Chikungunya virus)
- Fab chk-265 (Complex from Homo sapiens)
- E2 glycoprotein (38 kDa, Protein from Chikungunya virus)
- E1 glycoprotein (42 kDa, Protein from Chikungunya virus)
- Fab chk-265 heavy chain (23 kDa, Protein from Homo sapiens)
- Chikungunya virus-fab complex (Complex)