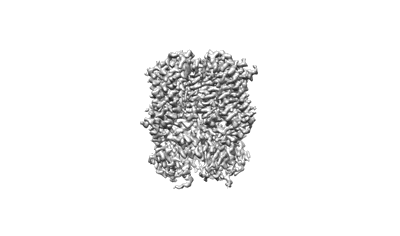
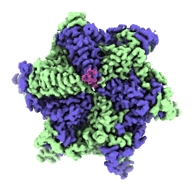
Cryo-EM structure of human HCN3 channel with cAMP
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 8io0
Q-score: 0.42
Cryo-EM structure of human HCN3 channel in apo state
Resolution: 2.72 Å
EM Method: Single-particle
Fitted PDBs: 8inz
Q-score: 0.526
Yu B, Lu Q, Li J, Cheng X, Hu H, Li Y, Che T, Hua Y, Jiang H, Zhang Y, Xian C, Yang T, Fu Y, Chen Y, Nan W, McCormick PJ, Xiong B, Duan J, Zeng B, Li Y, Fu Y, Zhang J
J Biol Chem (2024) 300 pp. 107288-107288 [ Pubmed: 38636662 DOI: doi:10.1016/j.jbc.2024.107288 ]
- 4-[[(2~{s},4~{a}~{r},6~{s},8~{a}~{s})-6-[(4~{s},5~{r})-4-[(2~{s})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{h}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid (492 Da, Ligand)
- Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (86 kDa, Protein from Homo sapiens)
- Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Complex from Homo sapiens)
Cryo-EM of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to nucleosome
Wang F, Feng X, He Q, Li H, Li H
J Biol Chem (2023) 299 pp. 102852-102852 [ Pubmed: 36592926 DOI: doi:10.1016/j.jbc.2022.102852 ]
- Yta7 and nucleosome complex (930 kDa, Complex from Saccharomyces cerevisiae)
- Yta7 (Protein from Saccharomyces cerevisiae)
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 7yhs
Q-score: 0.404
Gao Z, Zhang L, Ge Z, Wang H, Yue Y, Jiang Z, Wang X, Xu C, Zhang Y, Yang M, Feng Y
J Biol Chem (2022) 298 pp. 102575-102575 [ Pubmed: 36209819 DOI: doi:10.1016/j.jbc.2022.102575 ]
- Dna (39-mer) (16 kDa, DNA from Pseudomonas aeruginosa)
- Dna (5'-d(p*ap*gp*cp*ap*gp*cp*tp*gp*cp*ap*cp*c)-3') (16 kDa, DNA from Pseudomonas aeruginosa)
- Dna (Complex)
- Csy-acrif4 (Complex from Pseudomonas aeruginosa)
- Csy4 (21 kDa, Protein from Pseudomonas aeruginosa)
- Type i-f crispr-associated protein csy1 (49 kDa, Protein from Pseudomonas aeruginosa)
- Rna (Complex from Pseudomonas aeruginosa)
- Acrif4 (11 kDa, Protein from Pseudomonas aeruginosa)
- Rna (58-mer) (19 kDa, RNA from Pseudomonas aeruginosa)
- Crispr-associated protein csy3 (37 kDa, Protein from Pseudomonas aeruginosa)
- Crispr type i-f/ypest-associated protein csy2 (36 kDa, Protein from Pseudomonas aeruginosa)
- Csy-acrif4-dsdna (Complex from Pseudomonas aeruginosa)
Cryo-EM structure of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to ADP
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 7uqi
Q-score: 0.369
Wang F, Feng X, He Q, Li H, Li H
J Biol Chem (2022) 299 pp. 102852-102852 [ Pubmed: 36592926 DOI: doi:10.1016/j.jbc.2022.102852 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Atpase histone chaperone yta7 (162 kDa, Protein from Saccharomyces cerevisiae)
- Complex of yta7 with adp (930 kDa, Complex from Saccharomyces cerevisiae)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7uqj
Q-score: 0.362
Wang F, Feng X, He Q, Li H, Li H
J Biol Chem (2022) 299 pp. 102852-102852 [ Pubmed: 36592926 DOI: doi:10.1016/j.jbc.2022.102852 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
- Histone h3 (2 kDa, Protein from Saccharomyces cerevisiae)
- Magnesium ion (24 Da, Ligand)
- Atpase histone chaperone yta7 (162 kDa, Protein from Saccharomyces cerevisiae)
- Complex of yta7 with h3n tail (930 kDa, Complex from Saccharomyces cerevisiae)
Cryo-EM structure of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to ADP
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7uqk
Q-score: 0.417
Wang F, Feng X, He Q, Li H, Li H
J Biol Chem (2022) 299 pp. 102852-102852 [ Pubmed: 36592926 DOI: doi:10.1016/j.jbc.2022.102852 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Atpase histone chaperone yta7 (162 kDa, Protein from Saccharomyces cerevisiae)
- Complex of yta7 with adp (930 kDa, Complex from Saccharomyces cerevisiae)
the apo structure of human mTORC2 complex
Resolution: 3.28 Å
EM Method: Single-particle
Fitted PDBs: 7tzo
Q-score: 0.353
Yu Z, Chen J, Takagi E, Wang F, Saha B, Liu X, Joubert LM, Gleason CE, Jin M, Li C, Nowotny C, Agard D, Cheng Y, Pearce D
J Biol Chem (2022) 298 pp. 102288-102288 [ DOI: doi:10.1016/j.jbc.2022.102288 Pubmed: 35926713 ]
- Serine/threonine-protein kinase mtor (302 kDa, Protein from Homo sapiens)
- Target of rapamycin complex subunit lst8 (37 kDa, Protein from Homo sapiens)
- Target of rapamycin complex 2 subunit mapkap1 (60 kDa, Protein from Homo sapiens)
- Rapamycin-insensitive companion of mtor (193 kDa, Protein from Homo sapiens)
- Apostate of mtorc2 complex, composed of mtor, rictor, mlst8 and msin1 (Complex from Homo sapiens)
CryoEM structure of full length mouse TRPML2
Resolution: 3.18 Å
EM Method: Single-particle
Fitted PDBs: 7dys
Q-score: 0.438
Song X, Li J, Tian M, Zhu H, Hu X, Zhang Y, Cao Y, Ye H, McCormick PJ, Zeng B, Fu Y, Duan J, Zhang J
J Biol Chem (2022) 298 pp. 101487-101487 [ Pubmed: 34915027 DOI: doi:10.1016/j.jbc.2021.101487 ]
- Trpml2 (Cellular component from Mus musculus)
- Mucolipin-2 (60 kDa, Protein from Mus musculus)
CALHM1 open state with disordered CTH
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7dsc
Q-score: 0.372
Ren Y, Li Y, Wang Y, Wen T, Lu X, Chang S, Zhang X, Shen Y, Yang X
J Biol Chem (2022) 298 pp. 101838-101838 [ DOI: doi:10.1016/j.jbc.2022.101838 Pubmed: 35339491 ]
- Calhm1 (Cellular component from Danio rerio)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Calcium homeostasis modulator 1 (41 kDa, Protein from Danio rerio)