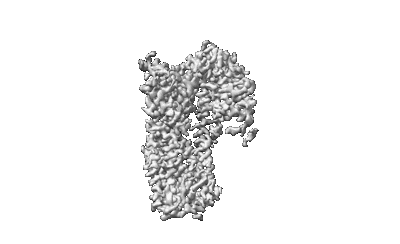
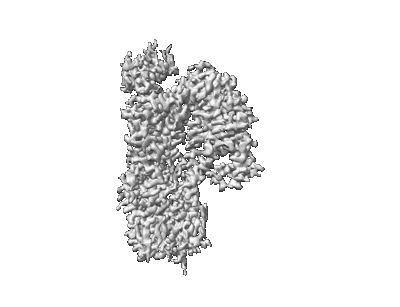
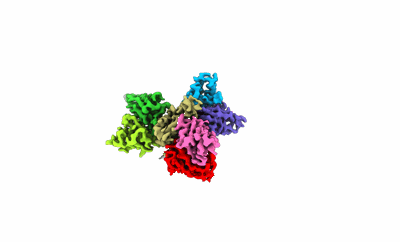
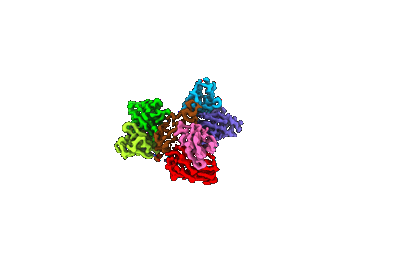
Cryo-EM structure of ATP13A2 in the E1-ATP state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8iek
Q-score: 0.377
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (128 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Cryo-em structure of atp13a2 in the e1-atp state (Complex from Homo sapiens)
Cryo-EM structure of ATP13A2 in the E1-like state
Resolution: 5.65 Å
EM Method: Single-particle
Fitted PDBs: 8iel
Q-score: 0.229
Cryo-EM structure of ATP13A2 in the E2P state
Resolution: 3.35 Å
EM Method: Single-particle
Fitted PDBs: 8iem
Q-score: 0.262
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (Complex from Homo sapiens)
- Polyamine-transporting atpase 13a2 (123 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Spermine (202 Da, Ligand)
- Beryllium trifluoride ion (66 Da, Ligand)
Cryo-EM structure of ATP13A2 in the E2-Pi state
Resolution: 3.25 Å
EM Method: Single-particle
Fitted PDBs: 8ien
Q-score: -0.047
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (128 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Tetrafluoroaluminate ion (102 Da, Ligand)
- Spermine (202 Da, Ligand)
- Cryo-em structure of atp13a2 in the e2-pi state (Complex from Homo sapiens)
Cryo-EM structure of ATP13A2 in the nominal E1P state
Resolution: 3.78 Å
EM Method: Single-particle
Fitted PDBs: 8ieo
Q-score: 0.312
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (128 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Tetrafluoroaluminate ion (102 Da, Ligand)
- Spermine (202 Da, Ligand)
- Cryo-em structure of atp13a2 in the nominal e1p state (Complex from Homo sapiens)
Cryo-EM structure of ATP13A2 in the putative of E2 state
Resolution: 4.87 Å
EM Method: Single-particle
Fitted PDBs: 8ier
Q-score: 0.162
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (128 kDa, Protein from Homo sapiens)
- Spermine (202 Da, Ligand)
- Cryo-em structure of atp13a2 in the putative of e2 state (Complex from Homo sapiens)
Cryo-EM structure of ATP13A2 in the E1P-ADP state
Resolution: 3.73 Å
EM Method: Single-particle
Fitted PDBs: 8ies
Q-score: 0.308
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Magnesium ion (24 Da, Ligand)
- Tetrafluoroaluminate ion (102 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Polyamine-transporting atpase 13a2 (125 kDa, Protein from Homo sapiens)
- Cryo-em structure of atp13a2 in the e1p-adp state (Complex from Homo sapiens)
Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17
Resolution: 3.48 Å
EM Method: Single-particle
Fitted PDBs: 7x7t
Q-score: 0.504
Xiong H, Sun H, Wang S, Yuan L, Liu L, Zhu Y, Zhang J, Huang Y, Qi R, Jiang Y, Ma J, Zhou M, Ma Y, Fu R, Yan S, Yue M, Wu Y, Wei M, Wang Y, Li T, Wang Y, Zheng Z, Yu H, Cheng T, Li S, Yuan Q, Zhang J, Guan Y, Zheng Q, Zhang T, Xia N
PNAS (2022) 119 pp. e2204256119-e2204256119 [ DOI: doi:10.1073/pnas.2204256119 Pubmed: 35972965 ]
- X17 heavy chain (13 kDa, Protein from Mus musculus)
- X01 light chain (11 kDa, Protein from Mus musculus)
- Nabs x01, x10 and x17 (Complex from Mus musculus)
- X10 light chain (12 kDa, Protein from Mus musculus)
- X17 light chain (11 kDa, Protein from Mus musculus)
- X10 heavy chain (13 kDa, Protein from Mus musculus)
- Sars-cov-2 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- X01 heavy chain (13 kDa, Protein from Mus musculus)
- Cryo-em structure of sars-cov-2 spike protein in complex with three nabs x01, x10 and x17 (Complex)
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Resolution: 3.77 Å
EM Method: Single-particle
Fitted PDBs: 7x7u
Q-score: 0.49
Xiong H, Sun H, Wang S, Yuan L, Liu L, Zhu Y, Zhang J, Huang Y, Qi R, Jiang Y, Ma J, Zhou M, Ma Y, Fu R, Yan S, Yue M, Wu Y, Wei M, Wang Y, Li T, Wang Y, Zheng Z, Yu H, Cheng T, Li S, Yuan Q, Zhang J, Guan Y, Zheng Q, Zhang T, Xia N
PNAS (2022) 119 pp. e2204256119-e2204256119 [ DOI: doi:10.1073/pnas.2204256119 Pubmed: 35972965 ]
- Spike protein s1 (23 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- X17 heavy chain (13 kDa, Protein from Mus musculus)
- X01 light chain (11 kDa, Protein from Mus musculus)
- Nabs x01, x10 and x17 (Complex from Mus musculus)
- X10 light chain (12 kDa, Protein from Mus musculus)
- Cryo-em structure of sars-cov-2 delta variant spike protein in complex with three nabs x01, x10 and x17 (Complex)
- X17 light chain (11 kDa, Protein from Mus musculus)
- X10 heavy chain (13 kDa, Protein from Mus musculus)
- Sars-cov-2 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- X01 heavy chain (13 kDa, Protein from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17
Resolution: 3.83 Å
EM Method: Single-particle
Fitted PDBs: 7x7v
Q-score: 0.484
Xiong H, Sun H, Wang S, Yuan L, Liu L, Zhu Y, Zhang J, Huang Y, Qi R, Jiang Y, Ma J, Zhou M, Ma Y, Fu R, Yan S, Yue M, Wu Y, Wei M, Wang Y, Li T, Wang Y, Zheng Z, Yu H, Cheng T, Li S, Yuan Q, Zhang J, Guan Y, Zheng Q, Zhang T, Xia N
PNAS (2022) 119 pp. e2204256119-e2204256119 [ DOI: doi:10.1073/pnas.2204256119 Pubmed: 35972965 ]
- X17 heavy chain (13 kDa, Protein from Mus musculus)
- X01 light chain (11 kDa, Protein from Mus musculus)
- Nabs x01, x10 and x17 (Complex from Mus musculus)
- X10 light chain (12 kDa, Protein from Mus musculus)
- Spike protein s1 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- X17 light chain (11 kDa, Protein from Mus musculus)
- X10 heavy chain (13 kDa, Protein from Mus musculus)
- Sars-cov spike protein (Complex from Severe acute respiratory syndrome coronavirus)
- Cryo-em structure of sars-cov spike protein in complex with three nabs x01, x10 and x17 (Complex)
- X01 heavy chain (13 kDa, Protein from Mus musculus)