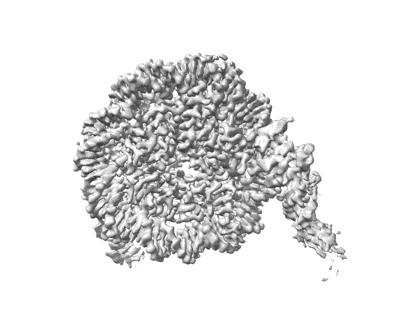
Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5
Resolution: 2.58 Å
EM Method: Single-particle
Fitted PDBs: 8pki
Q-score: 0.572
Kobayashi W, Sappler AH, Bollschweiler D, Kummecke M, Basquin J, Arslantas EN, Ruangroengkulrith S, Hornberger R, Duderstadt K, Tachibana K
Nat Struct Mol Biol (2024) 31 pp. 757-766 [ DOI: doi:10.1038/s41594-024-01239-0 Pubmed: 38409506 ]
- Histone h3.3 (15 kDa, Protein from Mus musculus)
- Dna (47 kDa, DNA from synthetic construct)
- Histone h2b type 1-c/e/g (13 kDa, Protein from Mus musculus)
- Dna (46 kDa, DNA from synthetic construct)
- Zinc ion (65 Da, Ligand)
- Histone h4 (11 kDa, Protein from Mus musculus)
- Nuclear receptor subfamily 5 group a member 2 (10 kDa, Protein from Homo sapiens)
- Cryo-em structure of the nucleosome containing nr5a2 motif at shl+5.5 (Complex from Mus musculus)
- Histone h2a (14 kDa, Protein from Mus musculus)
Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8pkj
Q-score: 0.6
Kobayashi W, Sappler AH, Bollschweiler D, Kummecke M, Basquin J, Arslantas EN, Ruangroengkulrith S, Hornberger R, Duderstadt K, Tachibana K
Nat Struct Mol Biol (2024) 31 pp. 757-766 [ DOI: doi:10.1038/s41594-024-01239-0 Pubmed: 38409506 ]
- Dna (47 kDa, DNA from synthetic construct)
- Histone h3 (fragment) (15 kDa, Protein from Mus musculus)
- Histone h2b (13 kDa, Protein from Mus musculus)
- Dna (46 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Mus musculus)
- Cryo-em structure of the nucleosome containing nr5a2 motif at shl+5.5 (Complex from Mus musculus)
- Histone h2a (14 kDa, Protein from Mus musculus)
CryoEM micrographs collected on a RAD51-ATP-dsDNA filament sample
Appleby R, Bollschweiler D, Chirgadze DY, Joudeh L, Pellegrini L
iScience (2023) 26 [ Pubmed: 37216117 DOI: 10.1016/j.isci.2023.106689 ]
Image sets:
CryoEM micrographs collected on a RAD51-ADP filament sample
Appleby R, Bollschweiler D, Chirgadze DY, Joudeh L, Pellegrini L
iScience (2023) 26 [ Pubmed: 37216117 DOI: 10.1016/j.isci.2023.106689 ]
Image sets:
CryoEM structure of the presynaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+
Resolution: 3.8 Å
EM Method: Helical reconstruction
Fitted PDBs: 8bq2
Q-score: 0.457
Appleby R, Bollschweiler D, Chirgadze DY, Joudeh L, Pellegrini L
iScience (2023) 26 pp. 106689-106689 [ Pubmed: 37216117 DOI: doi:10.1016/j.isci.2023.106689 ]
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Dna (30-mer) (9 kDa, DNA from Homo sapiens)
- Nucleoprotein filament of human rad51 bound to single-stranded dna 60mer in the presence of atp and ca2+ (Complex from Homo sapiens)
- Dna repair protein rad51 homolog 1 (37 kDa, Protein from Homo sapiens)
- Calcium ion (40 Da, Ligand)
CryoEM structure of the post-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8br2
Q-score: 0.573
Appleby R, Bollschweiler D, Chirgadze DY, Joudeh L, Pellegrini L
iScience (2023) 26 pp. 106689-106689 [ Pubmed: 37216117 DOI: doi:10.1016/j.isci.2023.106689 ]
- Dna (5'-d(p*tp*gp*gp*ap*gp*gp*tp*gp*cp*ap*tp*cp*gp*ap*gp*cp*tp*cp*gp*c)-3') (6 kDa, DNA from Homo sapiens)
- Dna (5'-d(p*gp*cp*gp*ap*gp*cp*tp*cp*gp*ap*tp*gp*cp*ap*cp*cp*tp*cp*cp*a)-3') (6 kDa, DNA from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Water (18 Da, Ligand)
- Post-synaptic rad51 nucleoprotein filament in the presence of atp and ca2+ (Complex from Homo sapiens)
- Dna repair protein rad51 homolog 1 (37 kDa, Protein from Homo sapiens)
- Calcium ion (40 Da, Ligand)
CryoEM structure of the RAD51 nucleoprotein filament in the presence of ADP and Ca2+
Resolution: 3.6 Å
EM Method: Helical reconstruction
Fitted PDBs: 8bsc
Q-score: 0.462
Appleby R, Bollschweiler D, Chirgadze DY, Joudeh L, Pellegrini L
iScience (2023) 26 pp. 106689-106689 [ Pubmed: 37216117 DOI: doi:10.1016/j.isci.2023.106689 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Filament of human rad51 bound to adp and ca2+ (Complex from Homo sapiens)
- Dna repair protein rad51 homolog 1 (37 kDa, Protein from Homo sapiens)
Cryo-EM structure of the Cetacean morbillivirus nucleoprotein-RNA complex
Resolution: 4.0 Å
EM Method: Helical reconstruction
Fitted PDBs: 7oi3
Q-score: 0.31
Zinzula L, Beck F, Klumpe S, Bohn S, Pfeifer G, Bollschweiler D, Nagy I, Plitzko JM, Baumeister W
J Struct Biol (2021) 213 pp. 107750-107750 [ Pubmed: 34089875 DOI: doi:10.1016/j.jsb.2021.107750 ]
- Poly-a 6-mer (Complex from Morbillivirus sp.)
- Cetacean morbillivirus nucleoprotein (46 kDa, Protein from Morbillivirus sp.)
- Poly-a 6-mer (1 kDa, RNA from Morbillivirus sp.)
- Cetacean morbillivirus nucleoprotein-rna complex (Complex)
- Cetacean morbillivirus nucleoprotein (Complex from Morbillivirus sp.)
Lee JH, Bollschweiler D, Schafer T, Huber R
Sci Adv (2021) 7 [ DOI: doi:10.1126/sciadv.abd4413 Pubmed: 33523989 ]
- Histone deacetylase (Complex from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Hdac-pc-nuc (Complex)
- Dna (Complex from unidentified plasmid)
- Histone (Complex from Xenopus laevis)
Resolution: 3.11 Å
EM Method: Single-particle
Fitted PDBs: 6z6f
Q-score: 0.471
Lee JH, Bollschweiler D, Schafer T, Huber R
Sci Adv (2021) 7 [ DOI: doi:10.1126/sciadv.abd4413 Pubmed: 33523989 ]
- Histone deacetylase hda1 (74 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Hda1 complex subunit 2 (71 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Histone deacetylase hda1 (76 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Hdac-pc (Complex from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Zinc ion (65 Da, Ligand)
- Hda1 complex subunit 3,hda1 complex subunit 3 (63 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))