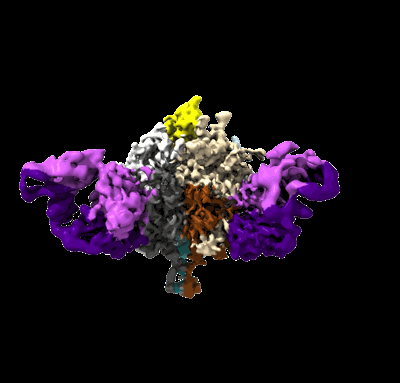
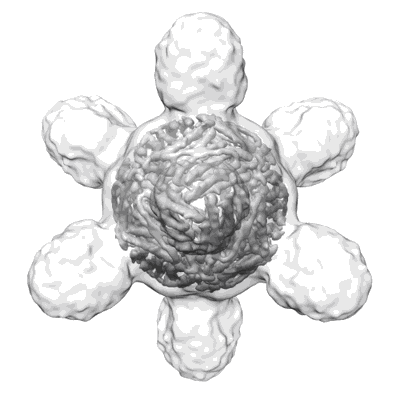
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 8t5c
Q-score: 0.301
Gorman J, Cheung CS, Duan Z, Ou L, Wang M, Chen X, Cheng C, Biju A, Sun Y, Wang P, Yang Y, Zhang B, Boyington JC, Bylund T, Charaf S, Chen SJ, Du H, Henry AR, Liu T, Sarfo EK, Schramm CA, Shen CH, Stephens T, Teng IT, Todd JP, Tsybovsky Y, Verardi R, Wang D, Wang S, Wang Z, Zheng CY, Zhou T, Douek DC, Mascola JR, Ho DD, Ho M, Kwong PD
Nat Commun (2024) 15 pp. 285-285 [ DOI: doi:10.1038/s41467-023-44534-y Pubmed: 38177144 ]
- Glycoprotein g1 (22 kDa, Protein from Lassa virus Josiah)
- D5 nanobody (13 kDa, Protein from Camelus bactrianus)
- Glycoprotein g2 (22 kDa, Protein from Lassa virus Josiah)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Lassa gpc trimer in complex with fab 8.11g and nanobody d5 (Complex from Lassa virus Josiah)
- 8.11g light chain (12 kDa, Protein from Homo sapiens)
- 8.11g heavy chain (13 kDa, Protein from Homo sapiens)
Lassa GPC trimer in complex with Fab GP23
Gorman J, Cheung CS, Duan Z, Ou L, Wang M, Chen X, Cheng C, Biju A, Sun Y, Wang P, Yang Y, Zhang B, Boyington JC, Bylund T, Charaf S, Chen SJ, Du H, Henry AR, Liu T, Sarfo EK, Schramm CA, Shen CH, Stephens T, Teng IT, Todd JP, Tsybovsky Y, Verardi R, Wang D, Wang S, Wang Z, Zheng CY, Zhou T, Douek DC, Mascola JR, Ho DD, Ho M, Kwong PD
Nat Commun (2024) 15 pp. 285-285 [ DOI: doi:10.1038/s41467-023-44534-y Pubmed: 38177144 ]
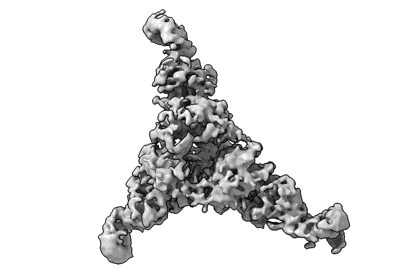
Ligand-free Lassa GPC Trimer with C3 Symmetry
Gorman J, Cheung CS, Duan Z, Ou L, Wang M, Chen X, Cheng C, Biju A, Sun Y, Wang P, Yang Y, Zhang B, Boyington JC, Bylund T, Charaf S, Chen SJ, Du H, Henry AR, Liu T, Sarfo EK, Schramm CA, Shen CH, Stephens T, Teng IT, Todd JP, Tsybovsky Y, Verardi R, Wang D, Wang S, Wang Z, Zheng CY, Zhou T, Douek DC, Mascola JR, Ho DD, Ho M, Kwong PD
Nat Commun (2024) 15 pp. 285-285 [ DOI: doi:10.1038/s41467-023-44534-y Pubmed: 38177144 ]
- Lassa gpc (Protein from Lassa virus Josiah)
- Lassa gpc trimer (Complex from Lassa virus Josiah)
Ligand-free Lassa GPC Trimer with C1 Symmetry
Gorman J, Cheung CS, Duan Z, Ou L, Wang M, Chen X, Cheng C, Biju A, Sun Y, Wang P, Yang Y, Zhang B, Boyington JC, Bylund T, Charaf S, Chen SJ, Du H, Henry AR, Liu T, Sarfo EK, Schramm CA, Shen CH, Stephens T, Teng IT, Todd JP, Tsybovsky Y, Verardi R, Wang D, Wang S, Wang Z, Zheng CY, Zhou T, Douek DC, Mascola JR, Ho DD, Ho M, Kwong PD
Nat Commun (2024) 15 pp. 285-285 [ DOI: doi:10.1038/s41467-023-44534-y Pubmed: 38177144 ]
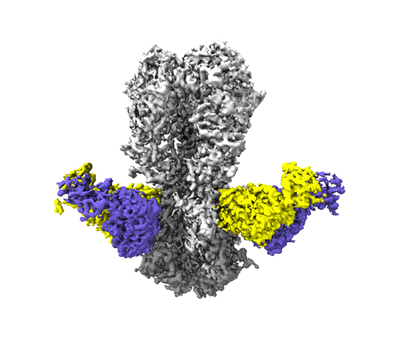
H10ssF: ferritin-based nanoparticle displaying H10 hemagglutinin stabilized stem epitopes
Moin SM, Boyington JC, Boyoglu-Barnum S, Gillespie RA, Cerutti G, Cheung CS, Cagigi A, Gallagher JR, Brand J, Prabhakaran M, Tsybovsky Y, Stephens T, Fisher BE, Creanga A, Ataca S, Rawi R, Corbett KS, Crank MC, Karlsson Hedestam GB, Gorman J, McDermott AB, Harris AK, Zhou T, Kwong PD, Shapiro L, Mascola JR, Graham BS, Kanekiyo M
Immunity (2022) 55 pp. 2405-2418.e7 [ Pubmed: 36356572 DOI: doi:10.1016/j.immuni.2022.10.015 ]
- H10ssf (Protein from Helicobacter pylori)
- H10ssf (1 MDa, Complex from Influenza A virus)
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 7l0l
Q-score: 0.555
Andrews SF, Raab JE, Gorman J, Gillespie RA, Cheung CSF, Rawi R, Cominsky LY, Boyington JC, Creanga A, Shen CH, Harris DR, Olia AS, Nazzari AF, Zhou T, Houser KV, Chen GL, Mascola JR, Graham BS, Kanekiyo M, Ledgerwood JE, Kwong PD, McDermott AB
Nat Med (2022) 28 pp. 373-382 [ DOI: doi:10.1038/s41591-021-01636-8 Pubmed: 35115707 ]
- Hemagglutinin ha1 chain (38 kDa, Protein from Influenza A virus (A/Canada/720/2005(H2N2)))
- 316-310-1b11 fab in complex with an h2 can05 ha trimer (Complex from Influenza A virus)
- Hemagglutinin ha2 chain (25 kDa, Protein from Influenza A virus (A/Canada/720/2005(H2N2)))
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 316-310-1b11 light chain (23 kDa, Protein from Homo sapiens)
- 316-310-1b11 heavy chain (25 kDa, Protein from Homo sapiens)
Resolution: 3.87 Å
EM Method: Single-particle
Fitted PDBs: 7mfg
Q-score: 0.411
Andrews SF, Raab JE, Gorman J, Gillespie RA, Cheung CSF, Rawi R, Cominsky LY, Boyington JC, Creanga A, Shen CH, Harris DR, Olia AS, Nazzari AF, Zhou T, Houser KV, Chen GL, Mascola JR, Graham BS, Kanekiyo M, Ledgerwood JE, Kwong PD, McDermott AB
Nat Med (2022) 28 pp. 373-382 [ DOI: doi:10.1038/s41591-021-01636-8 Pubmed: 35115707 ]
- 310-030-1d06 fab in complex with an h1 nc99 ha trimer (Complex from Influenza A virus)
- 310-030-1d06 heavy (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Hemagglutinin ha2 chain (25 kDa, Protein from Influenza A virus)
- Hemagglutinin ha1 chain (36 kDa, Protein from Influenza A virus)
- 310-030-1d06 light (11 kDa, Protein from Homo sapiens)
Resolution: 3.85 Å
EM Method: Single-particle
Fitted PDBs: 6xsk
Q-score: 0.406
Moin SM, Boyington JC, Boyoglu-Barnum S, Gillespie RA, Cerutti G, Cheung CS, Cagigi A, Gallagher JR, Brand J, Prabhakaran M, Tsybovsky Y, Stephens T, Fisher BE, Creanga A, Ataca S, Rawi R, Corbett KS, Crank MC, Karlsson Hedestam GB, Gorman J, McDermott AB, Harris AK, Zhou T, Kwong PD, Shapiro L, Mascola JR, Graham BS, Kanekiyo M
Immunity (2022) 55 pp. 2405-2418.e7 [ DOI: doi:10.1016/j.immuni.2022.10.015 Pubmed: 36356572 ]
- 789-203-3c12 fab heavy and light chains (Cellular component from Macaca mulatta)
- 789-203-3c12 fab heavy chain (49 kDa, Protein from Macaca mulatta)
- Hemagglutinin ha2 chain (25 kDa, Protein from Influenza A virus (A/Solomon Islands/3/2006(H1N1)))
- Hemagglutinin ha1 chain (38 kDa, Protein from Influenza A virus (A/Solomon Islands/3/2006(H1N1)))
- 789-203-3c12 fab light chain (23 kDa, Protein from Macaca mulatta)
- Influenza hemagglutinin (Cellular component from Influenza A virus (A/Solomon Islands/3/2006(H1N1)))
- Influenza hemagglutinin si06 trimer in complex with 789-203-3c12 fab (Cellular component)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
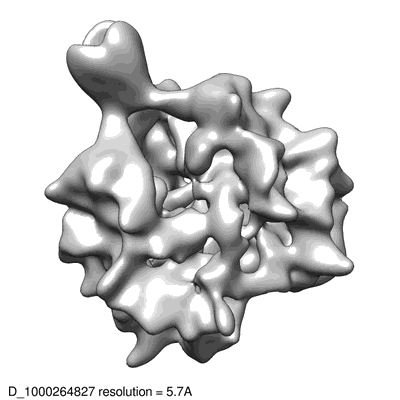
Cryo-EM structure of SARS-CoV-2 spike at pH 4.0
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schön A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879 [ Pubmed: 33271067 DOI: 10.1016/j.chom.2020.11.004 ]
Image sets:
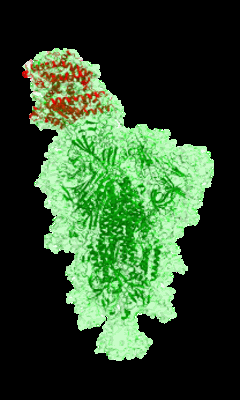
Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5
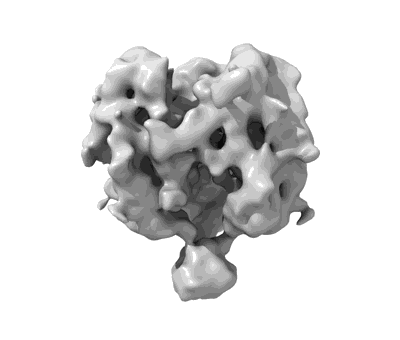
Resolution: 3.85 Å
EM Method: Single-particle
Fitted PDBs: 7kne
Q-score: 0.253
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Single ace2-bound sars-cov-2 trimer spike at ph 5.5 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Angiotensin-converting enzyme 2 (69 kDa, Protein from Homo sapiens)