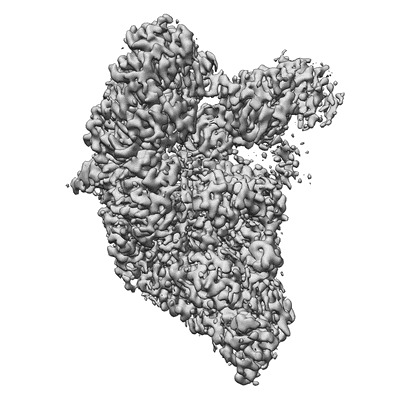
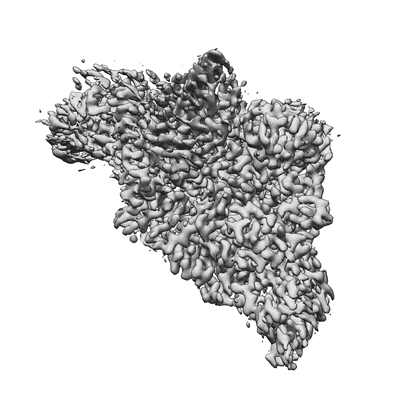
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8sq9
Q-score: 0.528
Small GI, Fedorova O, Olinares PDB, Chandanani J, Banerjee A, Choi YJ, Molina H, Chait BT, Darst SA, Campbell EA
Mol Cell (2023) 83 pp. 3921-3930.e7 [ DOI: doi:10.1016/j.molcel.2023.10.001 Pubmed: 37890482 ]
- Zinc ion (65 Da, Ligand)
- Template rna (17 kDa, RNA from Severe acute respiratory syndrome coronavirus)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
- Primer rna (11 kDa, RNA from Severe acute respiratory syndrome coronavirus)
- 5'-o-[(s)-hydroxy{[(s)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine (482 Da, Ligand)
- Non-structural protein 9 (12 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 replication-transcription complex bound to nsp9 and umpcpp, as a pre-catalytic nmpylation intermediate (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Resolution: 3.06 Å
EM Method: Single-particle
Fitted PDBs: 8sqj
Q-score: 0.511
Small GI, Fedorova O, Olinares PDB, Chandanani J, Banerjee A, Choi YJ, Molina H, Chait BT, Darst SA, Campbell EA
Mol Cell (2023) 83 pp. 3921-3930.e7 [ DOI: doi:10.1016/j.molcel.2023.10.001 Pubmed: 37890482 ]
- Zinc ion (65 Da, Ligand)
- Template rna (17 kDa, RNA from Severe acute respiratory syndrome coronavirus)
- Magnesium ion (24 Da, Ligand)
- Sars-cov-2 replication-transcription complex bound to rna-nsp9, as a noncatalytic rna-nsp9 binding mode (Complex from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
- Primer rna (11 kDa, RNA from Severe acute respiratory syndrome coronavirus)
- Non-structural protein 9 (12 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 5' utr (6 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 5'-o-[(r)-hydroxy(thiophosphonooxy)phosphoryl]guanosine (459 Da, Ligand)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Resolution: 3.01 Å
EM Method: Single-particle
Fitted PDBs: 8sqk
Q-score: 0.533
Small GI, Fedorova O, Olinares PDB, Chandanani J, Banerjee A, Choi YJ, Molina H, Chait BT, Darst SA, Campbell EA
Mol Cell (2023) 83 pp. 3921-3930.e7 [ DOI: doi:10.1016/j.molcel.2023.10.001 Pubmed: 37890482 ]
- Zinc ion (65 Da, Ligand)
- Sars-cov-2 replication-transcription complex bound to rna-nsp9 and gdp-betas, as a pre-catalytic dernaylation/mrna capping intermediate (Complex from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
- Rna-directed rna polymerase nsp12 (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Primer rna (10 kDa, RNA from Severe acute respiratory syndrome coronavirus)
- Non-structural protein 9 (12 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Template rna (11 kDa, RNA from Severe acute respiratory syndrome coronavirus)
- Sars-cov-2 5' utr (6 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 5'-o-[(r)-hydroxy(thiophosphonooxy)phosphoryl]guanosine (459 Da, Ligand)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Resolution: 3.18 Å
EM Method: Single-particle
Fitted PDBs: 8dy7
Q-score: 0.482
Lilic M, Holmes NA, Bush MJ, Marti AK, Widdick DA, Findlay KC, Choi YJ, Froom R, Koh S, Buttner MJ, Campbell EA
PNAS (2023) 120 pp. e2220785120-e2220785120 [ DOI: doi:10.1073/pnas.2220785120 Pubmed: 36888660 ]
- Zinc ion (65 Da, Ligand)
- Dna (78-mer) (30 kDa, DNA from Streptomyces venezuelae)
- Iron/sulfur cluster (351 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Rna polymerase sigma factor siga (56 kDa, Protein from Streptomyces venezuelae)
- Dna-directed rna polymerase subunit beta (130 kDa, Protein from Streptomyces venezuelae)
- Dna-directed rna polymerase subunit omega (9 kDa, Protein from Streptomyces venezuelae)
- Transcriptional regulator whib (10 kDa, Protein from Streptomyces venezuelae)
- Rna polymerase-binding protein rbpa (14 kDa, Protein from Streptomyces venezuelae)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Streptomyces venezuelae)
- Streptomyces venezuelae rnap transcription open promoter complex with whia and whib transcription factors (Complex from Streptomyces venezuelae)
- Probable cell division protein whia (37 kDa, Protein from Streptomyces venezuelae)
- Dna (71-mer) (30 kDa, DNA from Streptomyces venezuelae)
- Dna-directed rna polymerase subunit beta' (144 kDa, Protein from Streptomyces venezuelae)
Resolution: 3.12 Å
EM Method: Single-particle
Fitted PDBs: 8dy9
Q-score: 0.492
Lilic M, Holmes NA, Bush MJ, Marti AK, Widdick DA, Findlay KC, Choi YJ, Froom R, Koh S, Buttner MJ, Campbell EA
PNAS (2023) 120 pp. e2220785120-e2220785120 [ Pubmed: 36888660 DOI: doi:10.1073/pnas.2220785120 ]
- Magnesium ion (24 Da, Ligand)
- Rna polymerase-binding protein rbpa (14 kDa, Protein from Streptomyces venezuelae)
- Dna (38-mer) (15 kDa, DNA from Streptomyces venezuelae)
- Iron/sulfur cluster (351 Da, Ligand)
- Probable cell division protein whia (34 kDa, Protein from Streptomyces venezuelae)
- Rna polymerase sigma factor siga (56 kDa, Protein from Streptomyces venezuelae)
- Dna (38-mer) (15 kDa, DNA from Streptomyces venezuelae)
- Dna-directed rna polymerase subunit omega (9 kDa, Protein from Streptomyces venezuelae)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Streptomyces venezuelae ATCC 10712)
- Dna-directed rna polymerase subunit beta' (144 kDa, Protein from Streptomyces venezuelae)
- Zinc ion (65 Da, Ligand)
- Streptomyces venezuelae rnap unconstrained open promoter complex with whia and whib transcription factors (Complex from Streptomyces venezuelae)
- Dna-directed rna polymerase subunit beta (130 kDa, Protein from Streptomyces venezuelae)
- Transcriptional regulator whib (10 kDa, Protein from Streptomyces venezuelae)
Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7krn
Q-score: 0.476
Malone B, Chen J, Wang Q, Llewellyn E, Choi YJ, Olinares PDB, Cao X, Hernandez C, Eng ET, Chait BT, Shaw DE, Landick R, Darst SA, Campbell EA
PNAS (2021) 118 [ DOI: doi:10.1073/pnas.2102516118 Pubmed: 33883267 ]
- Zinc ion (65 Da, Ligand)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Chapso (631 Da, Ligand)
- Rna (43-mer) (17 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Aluminum fluoride (83 Da, Ligand)
- Helicase (67 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 backtracked complex bound to nsp13 helicase - nsp13(1)-btc (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (37-mer) (12 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Adenosine-5'-diphosphate (427 Da, Ligand)
Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-BTC
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7kro
Q-score: 0.416
Malone B, Chen J, Wang Q, Llewellyn E, Choi YJ, Olinares PDB, Cao X, Hernandez C, Eng ET, Chait BT, Shaw DE, Landick R, Darst SA, Campbell EA
PNAS (2021) 118 [ Pubmed: 33883267 DOI: doi:10.1073/pnas.2102516118 ]
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Aluminum fluoride (83 Da, Ligand)
- Rna (37-mer) (12 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Helicase (67 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (43-mer) (17 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Sars-cov-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-btc (Complex from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Chapso (631 Da, Ligand)
Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement)
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7krp
Q-score: 0.361
Malone B, Chen J, Wang Q, Llewellyn E, Choi YJ, Olinares PDB, Cao X, Hernandez C, Eng ET, Chait BT, Shaw DE, Landick R, Darst SA, Campbell EA
PNAS (2021) 118 [ Pubmed: 33883267 DOI: doi:10.1073/pnas.2102516118 ]
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Rna (37-mer) (12 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Rna (36-mer) (17 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Sars-cov-2 backtracked complex complex bound to nsp13 helicase - btc (local refinement) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Chapso (631 Da, Ligand)