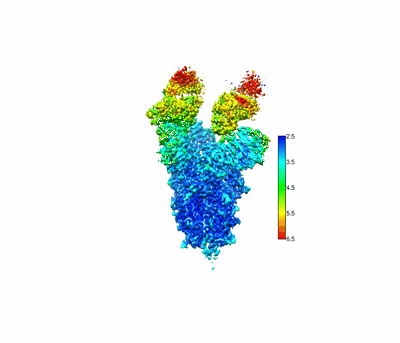
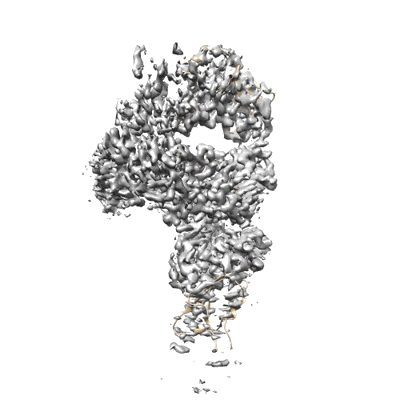SARS-CoV-2 Spike RBD (dimer) in complex with two 2S-1244 nanobodies
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 8jys
Q-score: 0.331
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8khc
Q-score: 0.351
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ DOI: doi:10.1016/j.celrep.2023.113653 Pubmed: 38175758 ]
- Light chain of 5817 fab (11 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron spike in complex with 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 fab (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
The interface structure of Omicron RBD binding to 5817 Fab
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8khd
Q-score: 0.393
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ DOI: doi:10.1016/j.celrep.2023.113653 Pubmed: 38175758 ]
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 (13 kDa, Protein from Homo sapiens)
- The interface structure of omicron rbd binding to 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Light chain of 5817 (11 kDa, Protein from Homo sapiens)
Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535
Resolution: 2.44 Å
EM Method: Single-particle
Fitted PDBs: 8h7l
Q-score: 0.377
Wang Y, Yan A, Song D, Duan M, Dong C, Chen J, Jiang Z, Gao Y, Rao M, Feng J, Zhang Z, Qi R, Ma X, Liu H, Yu B, Wang Q, Zong M, Jiao J, Xing P, Pan R, Li D, Xiao J, Sun J, Li Y, Zhang L, Shen Z, Sun B, Zhao Y, Zhang L, Dai J, Zhao J, Wang L, Dou C, Liu Z, Zhao J
Nat Commun (2024) 15 pp. 842-842 [ DOI: doi:10.1038/s41467-024-45050-3 Pubmed: 38287016 ]
- Sars-cov-2 ba.2 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Ba7535 fab light chain (23 kDa, Protein from Homo sapiens)
- Ba7535 (Complex from Homo sapiens)
- Cryo-em structure of sars-cov-2 ba.2 spike protein in complex with ba7535 (Complex)
- Ba7535 fab heavt chain (49 kDa, Protein from Homo sapiens)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of SARS-CoV-2 BA.2 RBD in complex with BA7535 fab (local refinement)
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 8h7z
Q-score: 0.461
Wang Y, Yan A, Song D, Duan M, Dong C, Chen J, Jiang Z, Gao Y, Rao M, Feng J, Zhang Z, Qi R, Ma X, Liu H, Yu B, Wang Q, Zong M, Jiao J, Xing P, Pan R, Li D, Xiao J, Sun J, Li Y, Zhang L, Shen Z, Sun B, Zhao Y, Zhang L, Dai J, Zhao J, Wang L, Dou C, Liu Z, Zhao J
Nat Commun (2024) 15 pp. 842-842 [ DOI: doi:10.1038/s41467-024-45050-3 Pubmed: 38287016 ]
- Spike glycoprotein (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ba7535 fab (49 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 ba.2 rbd in complex with ba7535 fab (local refinement) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Ba7535 fab (23 kDa, Protein from Homo sapiens)