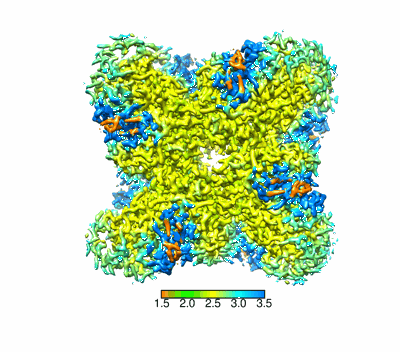
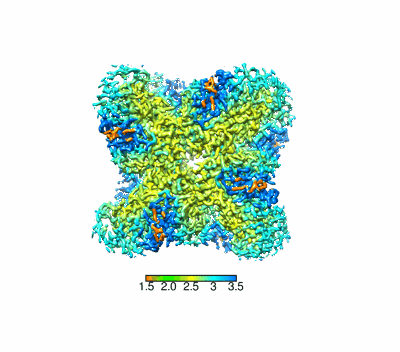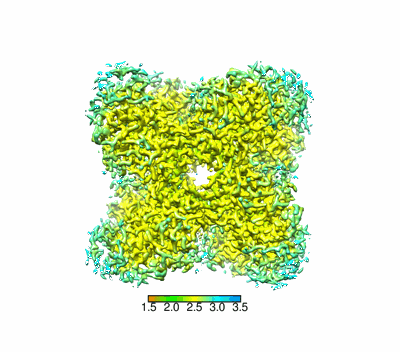Resolution: 2.37 Å
EM Method: Single-particle
Fitted PDBs: 8yak
Q-score: 0.652
Hu Y, Dai L, Xu Y, Niu D, Yang X, Xie Z, Shen P, Li X, Li H, Zhang L, Min J, Guo RT, Chen CC
Int J Biol Macromol (2024) 278 pp. 134831-134831 [ Pubmed: 39163957 DOI: doi:10.1016/j.ijbiomac.2024.134831 ]
- (2~{s})-2-[[(3~{r})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid (403 Da, Ligand)
- Imidazolonepropionase (43 kDa, Protein from Pseudoxanthomonas wuyuanensis)
- Pwadh-g88d-d344n (Complex from Pseudoxanthomonas wuyuanensis)
- Zinc ion (65 Da, Ligand)
Cryo-electron microscopic structure of an amide hydrolase from Pseudoxanthomonas wuyuanensis
Resolution: 2.33 Å
EM Method: Single-particle
Fitted PDBs: 8yag
Q-score: 0.665
Hu Y, Dai L, Xu Y, Niu D, Yang X, Xie Z, Shen P, Li X, Li H, Zhang L, Min J, Guo RT, Chen CC
Int J Biol Macromol (2024) 278 pp. 134831-134831 [ Pubmed: 39163957 DOI: doi:10.1016/j.ijbiomac.2024.134831 ]
- Pwadh (Complex from Pseudoxanthomonas wuyuanensis)
- Imidazolonepropionase (43 kDa, Protein from Pseudoxanthomonas wuyuanensis)
- Zinc ion (65 Da, Ligand)
Fab M2-7 complexed with SARS-Cov2 RBD and human ACE2
Resolution: 3.03 Å
EM Method: Single-particle
Fitted PDBs: 8xxw
Q-score: 0.484
Liu C, Xu S, Zheng Y, Xie Y, Xu K, Chai Y, Luo T, Dai L, Gao GF
Cell Rep (2024) 43 pp. 114235-114235 [ Pubmed: 38748880 DOI: doi:10.1016/j.celrep.2024.114235 ]
- Fab (Complex from Mus musculus)
- Ace2 (Complex from Homo sapiens)
- Spike protein s1 (20 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- M2-7-light chain (12 kDa, Protein from Mus musculus)
- Zinc ion (65 Da, Ligand)
- M2-7-heavy chain (13 kDa, Protein from Mus musculus)
- Fab m2-7 complexed with sars-cov2 rbd and human ace2 (Complex)
- Rbd (Complex from Severe acute respiratory syndrome coronavirus 2)
- Processed angiotensin-converting enzyme 2 (67 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3
Resolution: 2.71 Å
EM Method: Single-particle
Fitted PDBs: 8ihq
Q-score: 0.632
Dai L, Niu D, Huang JW, Li X, Shen P, Li H, Xie Z, Min J, Hu Y, Yang Y, Guo RT, Chen CC
J Hazard Mater (2023) 458 pp. 131836-131836 [ DOI: doi:10.1016/j.jhazmat.2023.131836 Pubmed: 37331057 ]
- Zinc ion (65 Da, Ligand)
- Amidohydrolase family protein (45 kDa, Protein from Stenotrophomonas acidaminiphila)
- Adh3 (Complex from Stenotrophomonas acidaminiphila)
Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8ihr
Q-score: 0.666
Dai L, Niu D, Huang JW, Li X, Shen P, Li H, Xie Z, Min J, Hu Y, Yang Y, Guo RT, Chen CC
J Hazard Mater (2023) 458 pp. 131836-131836 [ DOI: doi:10.1016/j.jhazmat.2023.131836 Pubmed: 37331057 ]
- Zinc ion (65 Da, Ligand)
- Amidohydrolase family protein (45 kDa, Protein from Stenotrophomonas acidaminiphila)
- Phenylalanine (165 Da, Ligand)
- Adh3 (Complex from Stenotrophomonas acidaminiphila)
Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8ihs
Q-score: 0.645
Dai L, Niu D, Huang JW, Li X, Shen P, Li H, Xie Z, Min J, Hu Y, Yang Y, Guo RT, Chen CC
J Hazard Mater (2023) 458 pp. 131836-131836 [ DOI: doi:10.1016/j.jhazmat.2023.131836 Pubmed: 37331057 ]
- Zinc ion (65 Da, Ligand)
- Amidohydrolase family protein (45 kDa, Protein from Stenotrophomonas acidaminiphila)
- (2~{s})-2-[[(3~{r})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid (403 Da, Ligand)
- Adh3 (Complex from Stenotrophomonas acidaminiphila)
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8j85
Q-score: 0.568
Dai L, Niu D, Huang JW, Li X, Shen P, Li H, Xie Z, Min J, Hu Y, Yang Y, Guo RT, Chen CC
J Hazard Mater (2023) 458 pp. 131836-131836 [ DOI: doi:10.1016/j.jhazmat.2023.131836 Pubmed: 37331057 ]
- Zinc ion (65 Da, Ligand)
- Amidohydrolase family protein (45 kDa, Protein from Stenotrophomonas acidaminiphila)
- (2~{s})-2-[[(3~{r})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid (403 Da, Ligand)
- Adh3 (Complex from Stenotrophomonas acidaminiphila)
Cryo-EM structure of SARS-CoV-2 prototype-Beta chimeric RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265- [ DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of SARS-CoV-2 Delta-Omicron chimeric RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265- [ DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265-2278.e14 [ Pubmed: 35568034 DOI: doi:10.1016/j.cell.2022.04.029 ]