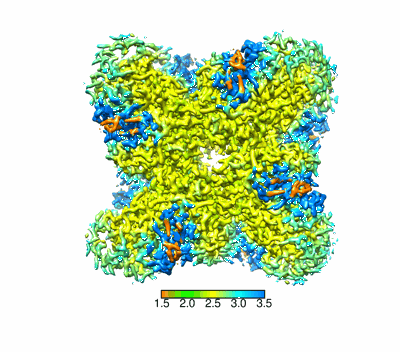
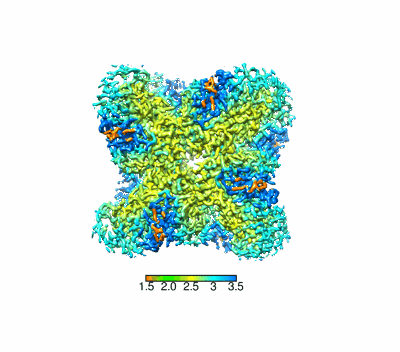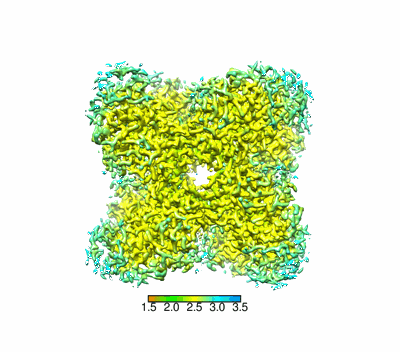Structural basis for the nucleosome binding and chromatin compaction by the linker histone H5
Structural basis for the nucleosome binding and chromatin compaction by the linker histone H5
Structural basis for the linker histone H5-nucleosome binding and chromatin compaction
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8xjv
Q-score: 0.424
Li W, Hu J, Song F, Yu J, Peng X, Zhang S, Wang L, Hu M, Liu JC, Wei Y, Xiao X, Li Y, Li D, Wang H, Zhou BR, Dai L, Mou Z, Zhou M, Zhang H, Zhou Z, Zhang H, Bai Y, Zhou JQ, Li W, Li G, Zhu P
Cell Res (2024) 34 pp. 707-724 [ DOI: doi:10.1038/s41422-024-01009-z Pubmed: 39103524 ]
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Histone h4 (Complex from Xenopus laevis)
- Histone h2b (Complex from Xenopus laevis)
- Linker histone h5 (Complex from Gallus gallus)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Histone h5 (21 kDa, Protein from Gallus gallus)
- Dna (660 kDa, DNA from synthetic construct)
- 12x177bp-h5 chromatin (Complex)
- Dna (651 kDa, DNA from synthetic construct)
- Histone h3.1 (Complex from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Widom 601 dna (Complex from synthetic construct)
- Histone h2a (Complex from Xenopus laevis)
Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3
Resolution: 2.71 Å
EM Method: Single-particle
Fitted PDBs: 8ihq
Q-score: 0.632
Dai L, Niu D, Huang JW, Li X, Shen P, Li H, Xie Z, Min J, Hu Y, Yang Y, Guo RT, Chen CC
J Hazard Mater (2023) 458 pp. 131836-131836 [ DOI: doi:10.1016/j.jhazmat.2023.131836 Pubmed: 37331057 ]
- Zinc ion (65 Da, Ligand)
- Amidohydrolase family protein (45 kDa, Protein from Stenotrophomonas acidaminiphila)
- Adh3 (Complex from Stenotrophomonas acidaminiphila)
Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8ihr
Q-score: 0.666
Dai L, Niu D, Huang JW, Li X, Shen P, Li H, Xie Z, Min J, Hu Y, Yang Y, Guo RT, Chen CC
J Hazard Mater (2023) 458 pp. 131836-131836 [ DOI: doi:10.1016/j.jhazmat.2023.131836 Pubmed: 37331057 ]
- Zinc ion (65 Da, Ligand)
- Amidohydrolase family protein (45 kDa, Protein from Stenotrophomonas acidaminiphila)
- Phenylalanine (165 Da, Ligand)
- Adh3 (Complex from Stenotrophomonas acidaminiphila)
Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8ihs
Q-score: 0.645
Dai L, Niu D, Huang JW, Li X, Shen P, Li H, Xie Z, Min J, Hu Y, Yang Y, Guo RT, Chen CC
J Hazard Mater (2023) 458 pp. 131836-131836 [ DOI: doi:10.1016/j.jhazmat.2023.131836 Pubmed: 37331057 ]
- Zinc ion (65 Da, Ligand)
- Amidohydrolase family protein (45 kDa, Protein from Stenotrophomonas acidaminiphila)
- (2~{s})-2-[[(3~{r})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid (403 Da, Ligand)
- Adh3 (Complex from Stenotrophomonas acidaminiphila)
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8j85
Q-score: 0.568
Dai L, Niu D, Huang JW, Li X, Shen P, Li H, Xie Z, Min J, Hu Y, Yang Y, Guo RT, Chen CC
J Hazard Mater (2023) 458 pp. 131836-131836 [ DOI: doi:10.1016/j.jhazmat.2023.131836 Pubmed: 37331057 ]
- Zinc ion (65 Da, Ligand)
- Amidohydrolase family protein (45 kDa, Protein from Stenotrophomonas acidaminiphila)
- (2~{s})-2-[[(3~{r})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid (403 Da, Ligand)
- Adh3 (Complex from Stenotrophomonas acidaminiphila)
Cryo-EM structure of SARS-CoV-2 prototype-Beta chimeric RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265- [ DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of SARS-CoV-2 Delta-Omicron chimeric RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265- [ DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265-2278.e14 [ Pubmed: 35568034 DOI: doi:10.1016/j.cell.2022.04.029 ]