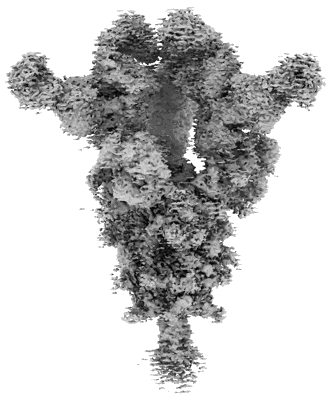
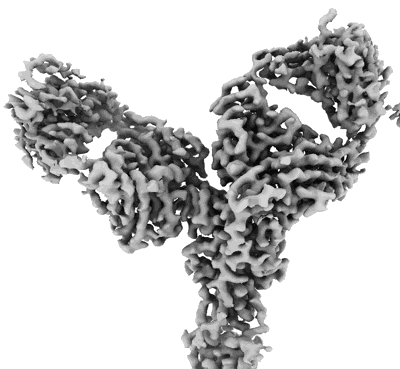
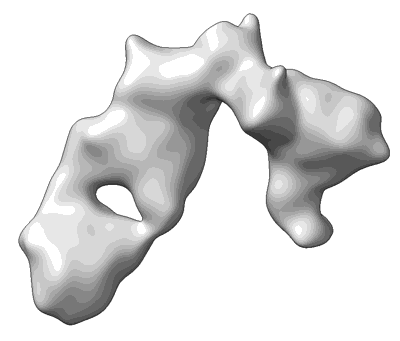
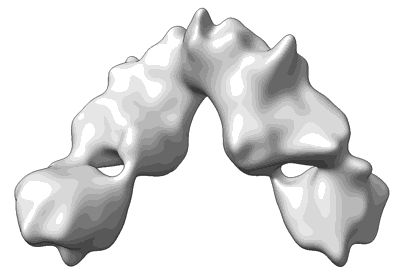
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 S-trimer in complex with fab L4.65 and L5.34
Resolution: 2.75 Å
EM Method: Single-particle
Fitted PDBs: 8hp9
Q-score: 0.368
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Spike protein s2' (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 omicron ba.2 s-trimer in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
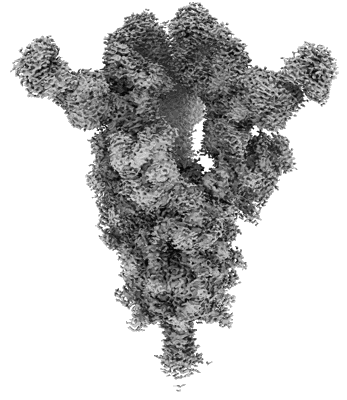
Cryo-EM structure of SARS-CoV-2 Omicron BA.4 S-trimer in complex with fab L4.65 and L5.34
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 8hpq
Q-score: 0.243
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Spike protein s2' (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cryo-em structure of sars-cov-2 omicron ba.4 s-trimer in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
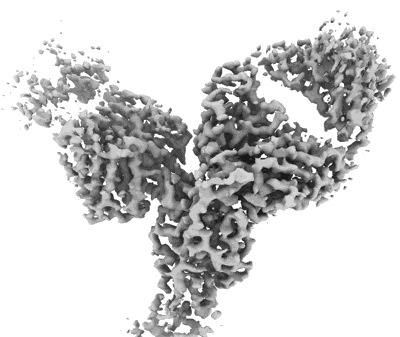
Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in complex with fab L4.65 and L5.34
Resolution: 2.65 Å
EM Method: Single-particle
Fitted PDBs: 8hpv
Q-score: 0.34
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Cryo-em structure of sars-cov-2 omicron prototype s-trimer in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike protein s2' (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
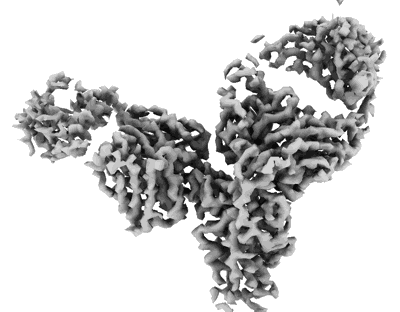
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with fab L4.65 and L5.34
Resolution: 2.34 Å
EM Method: Single-particle
Fitted PDBs: 8hpf
Q-score: 0.494
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 omicron ba.2 rbd in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Spike protein s2' (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34
Resolution: 2.56 Å
EM Method: Single-particle
Fitted PDBs: 8hpu
Q-score: 0.488
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Cryo-em structure of sars-cov-2 omicron ba.4 rbd in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- Spike protein s2' (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of SARS-CoV-2 Omicron Prototype RBD in complex with fab L4.65 and L5.34
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8hq7
Q-score: 0.528
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 omicron prototype rbd in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- Spike protein s1 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of SARS-CoV-2 prototype-Beta chimeric RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265- [ DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of SARS-CoV-2 Delta-Omicron chimeric RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265- [ DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265-2278.e14 [ Pubmed: 35568034 DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 5.5
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 6ilj
Q-score: 0.51
Zhao X, Zhang G, Liu S, Chen X, Peng R, Dai L, Qu X, Li S, Song H, Gao Z, Yuan P, Liu Z, Li C, Shang Z, Li Y, Zhang M, Qi J, Wang H, Du N, Wu Y, Bi Y, Gao S, Shi Y, Yan J, Zhang Y, Xie Z, Wei W, Gao GF
Cell (2019) 177 pp. 1553-1565.e16 [ DOI: doi:10.1016/j.cell.2019.04.035 Pubmed: 31104841 ]
- Complement decay-accelerating factor (21 kDa, Protein from Homo sapiens)
- Capsid protein vp3 (26 kDa, Protein from Echovirus E6)
- Capsid protein vp2 (28 kDa, Protein from Echovirus E6)
- Cryo-em structure of echovirus 6 complexed with its attachment receptor cd55 at ph 5.5 (Complex from Echovirus E6)
- Capsid protein vp1 (31 kDa, Protein from Echovirus E6)
- Sphingosine (299 Da, Ligand)
- Capsid protein vp4 (7 kDa, Protein from Echovirus E6)