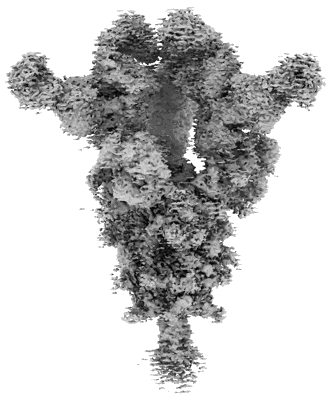
Resolution: 2.37 Å
EM Method: Single-particle
Fitted PDBs: 8yak
Q-score: 0.652
Hu Y, Dai L, Xu Y, Niu D, Yang X, Xie Z, Shen P, Li X, Li H, Zhang L, Min J, Guo RT, Chen CC
Int J Biol Macromol (2024) 278 pp. 134831-134831 [ Pubmed: 39163957 DOI: doi:10.1016/j.ijbiomac.2024.134831 ]
- (2~{s})-2-[[(3~{r})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid (403 Da, Ligand)
- Imidazolonepropionase (43 kDa, Protein from Pseudoxanthomonas wuyuanensis)
- Pwadh-g88d-d344n (Complex from Pseudoxanthomonas wuyuanensis)
- Zinc ion (65 Da, Ligand)
Cryo-electron microscopic structure of an amide hydrolase from Pseudoxanthomonas wuyuanensis
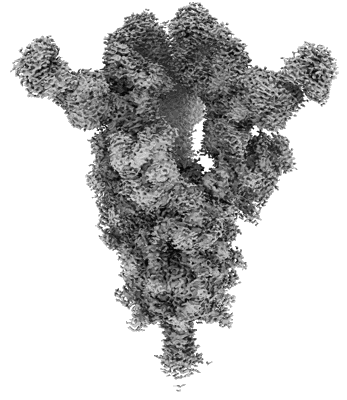
Resolution: 2.33 Å
EM Method: Single-particle
Fitted PDBs: 8yag
Q-score: 0.665
Hu Y, Dai L, Xu Y, Niu D, Yang X, Xie Z, Shen P, Li X, Li H, Zhang L, Min J, Guo RT, Chen CC
Int J Biol Macromol (2024) 278 pp. 134831-134831 [ Pubmed: 39163957 DOI: doi:10.1016/j.ijbiomac.2024.134831 ]
- Pwadh (Complex from Pseudoxanthomonas wuyuanensis)
- Imidazolonepropionase (43 kDa, Protein from Pseudoxanthomonas wuyuanensis)
- Zinc ion (65 Da, Ligand)
CryoEM structure of NaDC1 with Citrate
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8w6c
Q-score: 0.631
Chi X, Chen Y, Li Y, Dai L, Zhang Y, Shen Y, Chen Y, Shi T, Yang H, Wang Z, Yan R
Sci Adv (2024) 10 pp. eadl3685-eadl3685 [ DOI: doi:10.1126/sciadv.adl3685 Pubmed: 38552027 ]
- 1,2-diacyl-glycerol-3-sn-phosphate (704 Da, Ligand)
- Water (18 Da, Ligand)
- Cholesterol hemisuccinate (486 Da, Ligand)
- Solute carrier family 13 member 2 (65 kDa, Protein from Homo sapiens)
- Sodium ion (22 Da, Ligand)
- Citric acid (192 Da, Ligand)
- Structure of nadc1 in complex with citrate (Complex from Homo sapiens)
CryoEM structure of NaDC1 in apo state
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8w6d
Q-score: 0.662
Chi X, Chen Y, Li Y, Dai L, Zhang Y, Shen Y, Chen Y, Shi T, Yang H, Wang Z, Yan R
Sci Adv (2024) 10 pp. eadl3685-eadl3685 [ DOI: doi:10.1126/sciadv.adl3685 Pubmed: 38552027 ]
- 1,2-diacyl-glycerol-3-sn-phosphate (704 Da, Ligand)
- Cholesterol hemisuccinate (486 Da, Ligand)
- Structure of nadc1 in apo state (Complex from Homo sapiens)
- Solute carrier family 13 member 2 (65 kDa, Protein from Homo sapiens)
- Sodium ion (22 Da, Ligand)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8w6o
Q-score: 0.58
Chi X, Chen Y, Li Y, Dai L, Zhang Y, Shen Y, Chen Y, Shi T, Yang H, Wang Z, Yan R
Sci Adv (2024) 10 pp. eadl3685-eadl3685 [ DOI: doi:10.1126/sciadv.adl3685 Pubmed: 38552027 ]
- Cholesterol (386 Da, Ligand)
- Sodium ion (22 Da, Ligand)
- Solute carrier family 13 member 1 (67 kDa, Protein from Homo sapiens)
- Dimer of nas1 in in/in state (Complex from Homo sapiens)
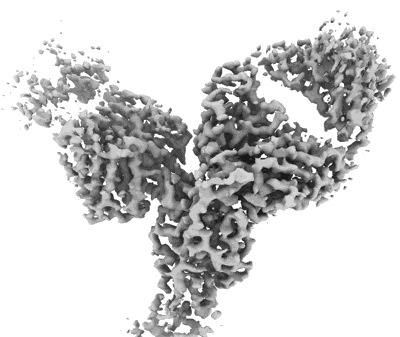
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 S-trimer in complex with fab L4.65 and L5.34
Resolution: 2.75 Å
EM Method: Single-particle
Fitted PDBs: 8hp9
Q-score: 0.368
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Spike protein s2' (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 omicron ba.2 s-trimer in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
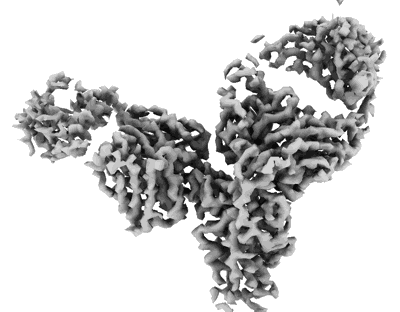
Cryo-EM structure of SARS-CoV-2 Omicron BA.4 S-trimer in complex with fab L4.65 and L5.34
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 8hpq
Q-score: 0.243
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Spike protein s2' (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cryo-em structure of sars-cov-2 omicron ba.4 s-trimer in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in complex with fab L4.65 and L5.34
Resolution: 2.65 Å
EM Method: Single-particle
Fitted PDBs: 8hpv
Q-score: 0.34
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Cryo-em structure of sars-cov-2 omicron prototype s-trimer in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike protein s2' (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with fab L4.65 and L5.34
Resolution: 2.34 Å
EM Method: Single-particle
Fitted PDBs: 8hpf
Q-score: 0.494
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 omicron ba.2 rbd in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Spike protein s2' (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34
Resolution: 2.56 Å
EM Method: Single-particle
Fitted PDBs: 8hpu
Q-score: 0.488
Guo S, Zheng Y, Gao Z, Duan M, Liu S, Du P, Xu X, Xu K, Zhao X, Chai Y, Wang P, Zhao Q, Gao GF, Dai L
Cell Discov (2023) 9 pp. 79-79 [ Pubmed: 37507370 DOI: doi:10.1038/s41421-023-00585-5 ]
- Cryo-em structure of sars-cov-2 omicron ba.4 rbd in complex with fab l4.65 and l5.34 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Fab l4.65 (24 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l5.34 (23 kDa, Protein from Homo sapiens)
- Fab l4.65 (23 kDa, Protein from Homo sapiens)
- Spike protein s2' (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)