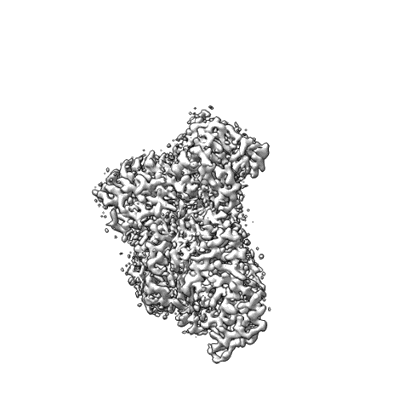
Cryo-EM structure of a Legionella effector complexed with actin and AMP
Resolution: 2.66 Å
EM Method: Single-particle
Fitted PDBs: 8jo3
Q-score: 0.618
Wang T, Song X, Tan J, Xian W, Zhou X, Yu M, Wang X, Xu Y, Wu T, Yuan K, Ran Y, Yang B, Fan G, Liu X, Zhou Y, Zhu Y
Nature (2024) 631 pp. 393-401 [ DOI: doi:10.1038/s41586-024-07573-z Pubmed: 38776962 ]
- Calcium ion (40 Da, Ligand)
- Substrate of the dot/icm secretion system (50 kDa, Protein from Legionella sainthelensi)
- Actin, alpha skeletal muscle (42 kDa, Protein from Oryctolagus cuniculus)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Cryo-em structure of a legionella effector complexed with actin and amp (Complex from Legionella sainthelensi)
- Adenosine monophosphate (347 Da, Ligand)
Cryo-EM structure of a Legionella effector complexed with actin and ATP
Resolution: 3.04 Å
EM Method: Single-particle
Fitted PDBs: 8jo4
Q-score: 0.51
Wang T, Song X, Tan J, Xian W, Zhou X, Yu M, Wang X, Xu Y, Wu T, Yuan K, Ran Y, Yang B, Fan G, Liu X, Zhou Y, Zhu Y
Nature (2024) 631 pp. 393-401 [ DOI: doi:10.1038/s41586-024-07573-z Pubmed: 38776962 ]
- Calcium ion (40 Da, Ligand)
- Cryo-em structure of a legionella effector complexed with actin and atp (Complex from Legionella sainthelensi)
- Substrate of the dot/icm secretion system (50 kDa, Protein from Legionella sainthelensi)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Actin, alpha skeletal muscle (42 kDa, Protein from Oryctolagus cuniculus)
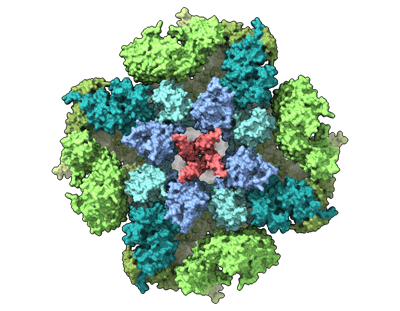
III2IV2 respiratory supercomplex from Saccharomyces cerevisiae with 4 bound UQ6
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8e7s
Q-score: 0.312
Hryc CF, Mallampalli VKPS, Bovshik EI, Azinas S, Fan G, Serysheva II, Sparagna GC, Baker ML, Mileykovskaya E, Dowhan W
Nat Commun (2023) 14 pp. 2783-2783 [ Pubmed: 37188665 DOI: doi:10.1038/s41467-023-38441-5 ]
- Cytochrome c oxidase subunit 4, mitochondrial (17 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 13, mitochondrial (15 kDa, Protein from Saccharomyces cerevisiae)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- Copper (ii) ion (63 Da, Ligand)
- 5-(3,7,11,15,19,23-hexamethyl-tetracosa-2,6,10,14,18,22-hexaenyl)-2,3-dimethoxy-6-methyl-benzene-1,4-diol (592 Da, Ligand)
- Cytochrome b-c1 complex subunit 9, mitochondrial (7 kDa, Protein from Saccharomyces cerevisiae)
- Dinuclear copper ion (127 Da, Ligand)
- Cytochrome b-c1 complex subunit 10, mitochondrial (8 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome b-c1 complex subunit 6, mitochondrial (17 kDa, Protein from Saccharomyces cerevisiae)
- Di-palmitoyl-3-sn-phosphatidylethanolamine (691 Da, Ligand)
- Cytochrome c oxidase subunit 6, mitochondrial (17 kDa, Protein from Saccharomyces cerevisiae)
- Iii2-iv2 mitochondrial respiratory supercomplex (Complex from Saccharomyces cerevisiae)
- Heme-a (852 Da, Ligand)
- Cytochrome b-c1 complex subunit 7, mitochondrial (14 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome b-c1 complex subunit 2, mitochondrial (40 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 3 (30 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 9, mitochondrial (6 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 1 (58 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c1, heme protein, mitochondrial (34 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 5a, mitochondrial (17 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome b-c1 complex subunit 1, mitochondrial (50 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 8, mitochondrial (8 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 12, mitochondrial (9 kDa, Protein from Saccharomyces cerevisiae)
- 1,2-diacyl-sn-glycero-3-phoshocholine (734 Da, Ligand)
- Cytochrome b-c1 complex subunit 8, mitochondrial (10 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 7, mitochondrial (6 kDa, Protein from Saccharomyces cerevisiae)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- (5s,11r)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-4,6,10,12,16-pentaoxa-5,11-diphosphaoctadec-1-yl pentadecanoate (634 Da, Ligand)
- Cytochrome c oxidase subunit 2 (28 kDa, Protein from Saccharomyces cerevisiae)
- Cardiolipin (1 kDa, Ligand)
- Cytochrome b (43 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome b-c1 complex subunit rieske, mitochondrial (23 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 26, mitochondrial (7 kDa, Protein from Saccharomyces cerevisiae)
III2IV respiratory supercomplex from Saccharomyces cerevisiae cardiolipin-lacking mutant
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8ec0
Q-score: 0.307
Hryc CF, Mallampalli VKPS, Bovshik EI, Azinas S, Fan G, Serysheva II, Sparagna GC, Baker ML, Mileykovskaya E, Dowhan W
Nat Commun (2023) 14 pp. 2783-2783 [ Pubmed: 37188665 DOI: doi:10.1038/s41467-023-38441-5 ]
- Cytochrome c oxidase subunit 13, mitochondrial (15 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome b-c1 complex subunit rieske, mitochondrial (23 kDa, Protein from Saccharomyces cerevisiae)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- (1s)-2-{[{[(2r)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl stearate (751 Da, Ligand)
- Copper (ii) ion (63 Da, Ligand)
- 5-(3,7,11,15,19,23-hexamethyl-tetracosa-2,6,10,14,18,22-hexaenyl)-2,3-dimethoxy-6-methyl-benzene-1,4-diol (592 Da, Ligand)
- Cytochrome b-c1 complex subunit 9, mitochondrial (7 kDa, Protein from Saccharomyces cerevisiae)
- Dinuclear copper ion (127 Da, Ligand)
- Iii2-iv mitochondrial respiratory supercomplex (Complex from Saccharomyces cerevisiae)
- Cytochrome b-c1 complex subunit 6, mitochondrial (17 kDa, Protein from Saccharomyces cerevisiae)
- Di-palmitoyl-3-sn-phosphatidylethanolamine (691 Da, Ligand)
- Cytochrome c oxidase subunit 6, mitochondrial (17 kDa, Protein from Saccharomyces cerevisiae)
- Heme-a (852 Da, Ligand)
- Cytochrome b-c1 complex subunit 7, mitochondrial (14 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome b-c1 complex subunit 2, mitochondrial (40 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 3 (30 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 9, mitochondrial (6 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 1 (58 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c1, heme protein, mitochondrial (34 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 5a, mitochondrial (17 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome b-c1 complex subunit 1, mitochondrial (50 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 8, mitochondrial (8 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 12, mitochondrial (9 kDa, Protein from Saccharomyces cerevisiae)
- 1,2-diacyl-sn-glycero-3-phoshocholine (734 Da, Ligand)
- Cytochrome b-c1 complex subunit 8, mitochondrial (10 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 7, mitochondrial (6 kDa, Protein from Saccharomyces cerevisiae)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Cytochrome c oxidase subunit 2 (28 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome b (43 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 4, mitochondrial (17 kDa, Protein from Saccharomyces cerevisiae)
- Cytochrome c oxidase subunit 26, mitochondrial (7 kDa, Protein from Saccharomyces cerevisiae)
Structure of the full-length IP3R1 channel determined at high Ca2+
Resolution: 3.26 Å
EM Method: Single-particle
Fitted PDBs: 8eaq
Q-score: 0.403
Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II
Nat Commun (2022) 13 pp. 6942-6942 [ Pubmed: 36376291 DOI: doi:10.1038/s41467-022-34574-1 ]
- Zinc ion (65 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- (9r,11s)-9-({[(1s)-1-hydroxyhexadecyl]oxy}methyl)-2,2-dimethyl-5,7,10-trioxa-2lambda~5~-aza-6lambda~5~-phosphaoctacosane-6,6,11-triol (767 Da, Ligand)
- Type 1 inositol 1,4,5-trisphosphate receptor tetrameric protein complex (1 MDa, Complex from Rattus norvegicus)
- Inositol 1,4,5-trisphosphate receptor type 1 (313 kDa, Protein from Rattus norvegicus)
Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8ear
Q-score: 0.35
Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II
Nat Commun (2022) 13 pp. 6942-6942 [ Pubmed: 36376291 DOI: doi:10.1038/s41467-022-34574-1 ]
- Zinc ion (65 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- (9r,11s)-9-({[(1s)-1-hydroxyhexadecyl]oxy}methyl)-2,2-dimethyl-5,7,10-trioxa-2lambda~5~-aza-6lambda~5~-phosphaoctacosane-6,6,11-triol (767 Da, Ligand)
- Type 1 inositol 1,4,5-trisphosphate receptor tetrameric protein complex (1 MDa, Complex from Rattus norvegicus)
- Inositol 1,4,5-trisphosphate receptor type 1 (313 kDa, Protein from Rattus norvegicus)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- D-myo-inositol-1,4,5-triphosphate (420 Da, Ligand)
Subnanometer in situ structure of the AcrAB-TolC efflux pump bound with MBX
Chen M, Shi X, Yu Z, Fan G, Serysheva II, Baker ML, Luisi BF, Ludtke SJ, Wang Z
Structure (2022) 30 pp. 107-113.e3 [ DOI: doi:10.1016/j.str.2021.08.008 Pubmed: 34506732 ]
- Acrab-tolc (Complex from Escherichia coli)
Structure of full-length IP3R1 channel reconstituted into lipid nanodisc in the apo-state
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7lhe
Q-score: 0.346
Baker MR, Fan G, Seryshev AB, Agosto MA, Baker ML, Serysheva II
Commun Biol (2021) 4 pp. 625- [ DOI: doi:10.1038/s42003-021-02156-4 ]
- Zinc ion (65 Da, Ligand)
- Type 1 inositol 1,4,5-trisphosphate receptor tetrameric protein complex (1 MDa, Complex from Rattus norvegicus)
- (9r,11s)-9-({[(1s)-1-hydroxyhexadecyl]oxy}methyl)-2,2-dimethyl-5,7,10-trioxa-2lambda~5~-aza-6lambda~5~-phosphaoctacosane-6,6,11-triol (767 Da, Ligand)
- Inositol 1,4,5-trisphosphate receptor type 1 (311 kDa, Protein from Rattus norvegicus)
Structure of full-length IP3R1 channel solubilized in LNMG lipid in the apo-state
Resolution: 2.96 Å
EM Method: Single-particle
Fitted PDBs: 7lhf
Q-score: 0.376
Baker MR, Fan G, Seryshev AB, Agosto MA, Baker ML, Serysheva II
Commun Biol (2021) 4 pp. 625- [ DOI: doi:10.1038/s42003-021-02156-4 ]
- Inositol 1,4,5-trisphosphate receptor type 1 (312 kDa, Protein from Rattus norvegicus)
- Zinc ion (65 Da, Ligand)
- Type 1 inositol 1,4,5-trisphosphate receptor tetrameric protein complex (1 MDa, Complex from Rattus norvegicus)
- (9r,11s)-9-({[(1s)-1-hydroxyhexadecyl]oxy}methyl)-2,2-dimethyl-5,7,10-trioxa-2lambda~5~-aza-6lambda~5~-phosphaoctacosane-6,6,11-triol (767 Da, Ligand)
Open state structure of the full-length TRPV2 cation channel with a resolved pore turret domain
Dosey TL, Wang Z, Fan G, Zhang Z, Serysheva II, Chiu W, Wensel TG
Nat. Struct. Mol. Biol. (2018) 26 pp. 40-49 [ DOI: 10.1038/s41594-018-0168-8 Pubmed: 30598551 ]
Image sets: