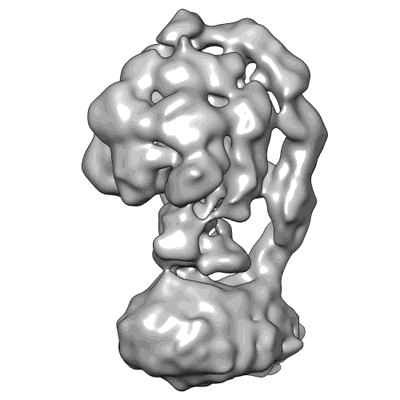
Resolution: 8.0 Å
EM Method: Single-particle
Fitted PDBs: 8ydy
Q-score: 0.084
Nakamura S, Tanimura Y, Nomura R, Suzuki H, Nishikawa K, Kamegawa A, Numoto N, Tanaka A, Kawabata S, Sakaguchi S, Emi A, Suzuki Y, Fujiyoshi Y
To Be Published
- Cespiace (4 kDa, Protein from synthetic construct)
- Sars-cov-2 spike ectodomain (hexapro, omicron ba.5 variant) in complex with cespiace (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (144 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Hexadecameric structure of an invertebrate gap junction channel
Oshima A, Matsuzawa T, Murata K, Tani K, Fujiyoshi Y
J Mol Biol (2016) 428 pp. 1227-1236 [ DOI: doi:10.1016/j.jmb.2016.02.011 Pubmed: 26883891 ]
- Innexin-6 (700 kDa, Protein from Caenorhabditis elegans)
- The n-terminal deleted c. elegans innexin-6 (700 kDa, Sample)
Hexadecameric structure of an invertebrate gap junction channel
Oshima A, Matsuzawa T, Murata K, Tani K, Fujiyoshi Y
J Mol Biol (2016) 428 pp. 1227-1236 [ DOI: doi:10.1016/j.jmb.2016.02.011 Pubmed: 26883891 ]
- Innexin-6 (700 kDa, Protein from Caenorhabditis elegans)
- The n-terminal deleted c. elegans innexin-6 (700 kDa, Sample)
Cryo-EM structure of bovine FoF1 ATP synthase
Hauer F, Gerle C, Fischer N, Oshima A, Shinzawa-Itoh K, Shimada S, Yokoyama K, Fujiyoshi Y, Stark H
Structure (2015) 23 pp. 1769-1775 [ Pubmed: 26278176 DOI: doi:10.1016/j.str.2015.06.029 ]
- Fof1 atp synthase (600 kDa, Protein from Bos taurus)
- Bovine mitochondrial fof1 atp synthase (600 kDa, Sample)
Cryo-EM structure of antagonist-bound E2P gastric H+,K+-ATPase (SCH.E2.AlF)
Resolution: 8.0 Å
EM Method: Electron Crystallography
Fitted PDBs: 4ux1
Q-score: 0.106
J Biol Chem (2014) 289 pp. 30590-30601 [ DOI: doi:10.1074/jbc.M114.584623 Pubmed: 25231997 ]
- H+,k+-atpase (110 kDa, Protein from Sus scrofa)
- Potassium-transporting atpase (Sample)
cryo-EM structure of the NavCt voltage-gated sodium channel
Resolution: 9.0 Å
EM Method: Electron Crystallography
Fitted PDBs: 4bgn
Q-score: 0.064
Tsai CJ, Tani K, Irie K, Hiroaki Y, Shimomura T, McMillan DG, Cook GM, Schertler GF, Fujiyoshi Y, Li XD
J Mol Biol (2013) 425 pp. 4074-4088 [ Pubmed: 23831224 DOI: doi:10.1016/j.jmb.2013.06.036 ]
Electron crystallographic studies on IP39 of Euglena gracilis with Fab
Suzuki H, Ito Y, Yamazaki Y, Mineta K, Uji M, Abe K, Tani K, Fujiyoshi Y, Tsukita S
Nat Commun (2013) 4 pp. 1766-1766 [ Pubmed: 23612307 DOI: doi:10.1038/ncomms2731 ]
- Alpha-type ip39 (39 kDa, Protein from Euglena gracilis)
- Euglena ip39 (39 kDa, Sample)
Cryo-EM structure of gastric H+,K+-ATPase with bound Rb+
Resolution: 8.0 Å
EM Method: Electron Crystallography
Fitted PDBs: 2yn9
Q-score: 0.059
Abe K, Tani K, Friedrich T, Fujiyoshi Y
PNAS (2012) 109 pp. 18401-18406 [ DOI: doi:10.1073/pnas.1212294109 Pubmed: 23091039 ]
- Potassium-transporting atpase (150 kDa, Protein from Sus scrofa)
- H+,k+-atpase bound with rb+ (150 kDa, Sample)
Cryo-EM structure of gastric H+,K+-ATPase with bound K+
Abe K, Tani K, Friedrich T, Fujiyoshi Y
PNAS (2012) 109 pp. 18401-18406 [ DOI: doi:10.1073/pnas.1212294109 Pubmed: 23091039 ]
- Potassium-transporting atpase (150 kDa, Protein from Sus scrofa)
- Pig gastric h,k-atpase (150 kDa, Sample)
A structure of Cx26M34Adel2-7 at 10 angstrom resolution
Resolution: 10.0 Å
EM Method: Electron Crystallography
Fitted PDBs: 3iz2
Q-score: 0.092
Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y
J Mol Biol (2011) 405 pp. 724-735 [ Pubmed: 21094651 DOI: doi:10.1016/j.jmb.2010.10.032 ]
- Gap junction (Cellular component from Homo sapiens)
- Human connexin26 (300 kDa, Sample)