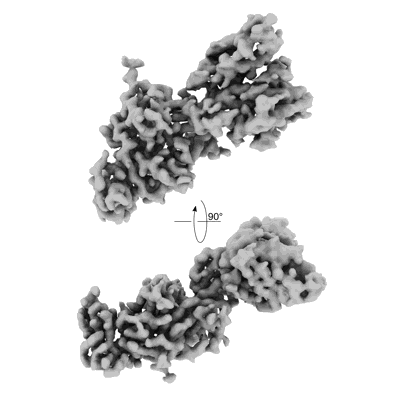SARS-CoV-2 Spike NTD in complex with neutralizing Fab SARS2-57 (local refinement)
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 7sww
Q-score: 0.328
Adams LJ, VanBlargan LA, Liu Z, Gilchuk P, Zhao H, Chen RE, Raju S, Whitener B, Shrihari S, Jethva PN, Gross ML, Crowe JE, Whelan SPJ, Diamond MS, Fremont DH
To Be Published
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars2-57 fv light chain (12 kDa, Protein from Mus musculus)
- Spike protein s1 (33 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars2-57 fv heavy chain (12 kDa, Protein from Mus musculus)
- Sars-cov-2 spike bound by sars2-57 fab (Complex)
SARS-CoV-2 Spike in complex with neutralizing Fab SARS2-57 (three down conformation)
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 7swx
Q-score: 0.446
Adams LJ, VanBlargan LA, Liu Z, Gilchuk P, Zhao H, Chen RE, Raju S, Whitener B, Shrihari S, Jethva PN, Gross ML, Crowe JE, Whelan SPJ, Diamond MS, Fremont DH
To Be Published
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars2-57 fv light chain (12 kDa, Protein from Mus musculus)
- Sars2-57 fv heavy chain (12 kDa, Protein from Mus musculus)
- Sars-cov-2 spike bound by sars2-57 fab (Complex)
- Spike glycoprotein (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 Spike in complex with neutralizing Fab SARS2-38 (three down conformation)
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7mkl
Q-score: 0.228
VanBlargan LA, Adams LJ, Liu Z, Chen RE, Gilchuk P, Raju S, Smith BK, Zhao H, Case JB, Winkler ES, Whitener BM, Droit L, Aziati ID, Bricker TL, Joshi A, Shi PY, Creanga A, Pegu A, Handley SA, Wang D, Boon ACM, Crowe Jr JE, Whelan SPJ, Fremont DH, Diamond MS
Immunity (2021) 54 pp. 2399-2416.e6 [ Pubmed: 34481543 DOI: doi:10.1016/j.immuni.2021.08.016 ]
- Sars2-38 fv heavy chain (12 kDa, Protein from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars2-38 fv light chain (11 kDa, Protein from Mus musculus)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike bound by sars2-38 antibody fab (Complex from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 Spike RBD in complex with neutralizing Fab SARS2-38 (local refinement)
Resolution: 3.16 Å
EM Method: Single-particle
Fitted PDBs: 7mkm
Q-score: 0.3
VanBlargan LA, Adams LJ, Liu Z, Chen RE, Gilchuk P, Raju S, Smith BK, Zhao H, Case JB, Winkler ES, Whitener BM, Droit L, Aziati ID, Bricker TL, Joshi A, Shi PY, Creanga A, Pegu A, Handley SA, Wang D, Boon ACM, Crowe Jr JE, Whelan SPJ, Fremont DH, Diamond MS
Immunity (2021) 54 pp. 2399-2416.e6 [ Pubmed: 34481543 DOI: doi:10.1016/j.immuni.2021.08.016 ]
- Sars2-38 fv heavy chain (12 kDa, Protein from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars2-38 fv light chain (11 kDa, Protein from Mus musculus)
- Spike protein s1 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike bound by sars2-38 antibody fab (Complex from Severe acute respiratory syndrome coronavirus 2)
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2082
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe Jr JE, Bloom JD
Cell Host Microbe (2021) 29 pp. 44-57.e9 [ Pubmed: 33259788 DOI: doi:10.1016/j.chom.2020.11.007 ]
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2479
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe Jr JE, Bloom JD
Cell Host Microbe (2021) 29 pp. 44-57.e9 [ Pubmed: 33259788 DOI: doi:10.1016/j.chom.2020.11.007 ]
Negative stain EM map of SARS-COV-2 spike protein open RBD (trimer) with Fab COV2-2096
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe Jr JE, Bloom JD
Cell Host Microbe (2021) 29 pp. 44-57.e9 [ Pubmed: 33259788 DOI: doi:10.1016/j.chom.2020.11.007 ]
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2832
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe Jr JE, Bloom JD
Cell Host Microbe (2021) 29 pp. 44-57.e9 [ Pubmed: 33259788 DOI: doi:10.1016/j.chom.2020.11.007 ]