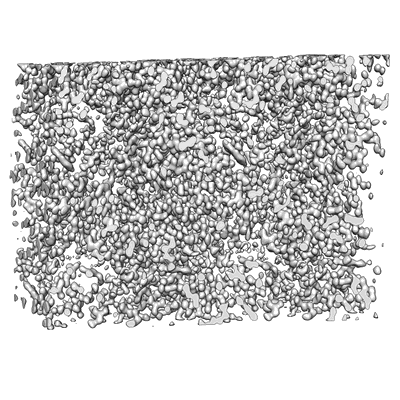
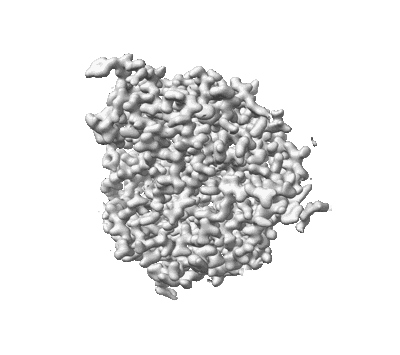
MicroED structure of triclinic lysozyme
Clabbers MTB, Martynowycz MW, Hattne J, Gonen T
J Struct Biol X (2022) 6 pp. 100078-100078 [ DOI: doi:10.1016/j.yjsbx.2022.100078 Pubmed: 36507068 ]
- Lysozyme c (14 kDa, Protein from Gallus gallus)
- Nitrate ion (62 Da, Ligand)
- Water (18 Da, Ligand)
- Lysozyme (14 kDa, Complex from Gallus gallus)
MicroED structure of proteinase K recorded on K2
Clabbers MTB, Martynowycz MW, Hattne J, Nannenga BL, Gonen T
J Struct Biol (2022) 214 pp. 107886-107886 [ Pubmed: 36044956 DOI: doi:10.1016/j.jsb.2022.107886 ]
- Proteinase k (28 kDa, Complex from Parengyodontium album)
- Calcium ion (40 Da, Ligand)
- Proteinase k (28 kDa, Protein from Parengyodontium album)
MicroED structure of proteinase K recorded on K3
Clabbers MTB, Martynowycz MW, Hattne J, Nannenga BL, Gonen T
J Struct Biol (2022) 214 pp. 107886-107886 [ Pubmed: 36044956 DOI: doi:10.1016/j.jsb.2022.107886 ]
- Proteinase k (28 kDa, Protein from Parengyodontium album)
- Proteinase k (28 kDa, Complex from Parengyodontium album)
- Calcium ion (40 Da, Ligand)
- Water (18 Da, Ligand)
MicroED structure of triclinic lysozyme recorded on K3
Clabbers MTB, Martynowycz MW, Hattne J, Nannenga BL, Gonen T
J Struct Biol (2022) 214 pp. 107886-107886 [ Pubmed: 36044956 DOI: doi:10.1016/j.jsb.2022.107886 ]
- Lysozyme c (16 kDa, Protein from Gallus gallus)
- Nitrate ion (62 Da, Ligand)
- Water (18 Da, Ligand)
- Lysozyme (14 kDa, Complex from Gallus gallus)
Ab initio structure of triclinic lysozyme from electron-counted MicroED data
Martynowycz MW, Clabbers MTB, Hattne J, Gonen T
Nat Methods (2022) 19 pp. 724-729 [ DOI: doi:10.1038/s41592-022-01485-4 Pubmed: 35637302 ]
- Lysozyme c (14 kDa, Protein from Gallus gallus)
- Nitrate ion (62 Da, Ligand)
- Water (18 Da, Ligand)
- Lysozyme (14 kDa, Complex from Gallus gallus)
Ab initio structure of proteinase K from electron-counted MicroED data
Martynowycz MW, Clabbers MTB, Hattne J, Gonen T
Nat Methods (2022) 19 pp. 724-729 [ DOI: doi:10.1038/s41592-022-01485-4 Pubmed: 35637302 ]
- Calcium ion (40 Da, Ligand)
- 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid (558 Da, Ligand)
- Proteinase k (28 kDa, Complex from Parengyodontium album)
- Proteinase k (28 kDa, Protein from Parengyodontium album)
- Water (18 Da, Ligand)
MicroED structure of proteinase K recorded on Falcon III
Resolution: 2.1 Å
EM Method: Electron Crystallography
Fitted PDBs: 6pu4
Q-score: 0.794
Hattne J, Martynowycz MW, Penczek PA, Gonen T
IUCrJ (2019) 6 pp. 921-926 [ Pubmed: 31576224 DOI: doi:10.1107/S2052252519010583 ]
- Calcium ion (40 Da, Ligand)
- Proteinase k (29 kDa, Complex from Parengyodontium album)
- Water (18 Da, Ligand)
- Proteinase k (28 kDa, Protein from Parengyodontium album)
MicroED structure of proteinase K recorded on CetaD
Resolution: 2.7 Å
EM Method: Electron Crystallography
Fitted PDBs: 6pu5
Q-score: 0.678
Hattne J, Martynowycz MW, Penczek PA, Gonen T
IUCrJ (2019) 6 pp. 921-926 [ Pubmed: 31576224 DOI: doi:10.1107/S2052252519010583 ]
- Water (18 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Proteinase k (28 kDa, Complex from Parengyodontium album)
- Proteinase k (28 kDa, Protein from Parengyodontium album)