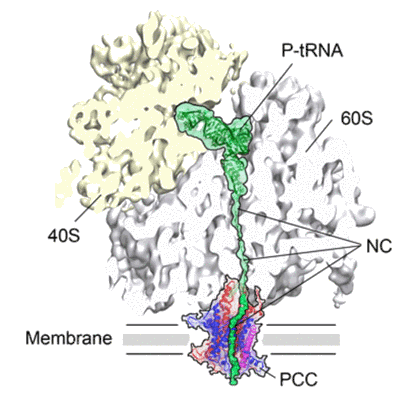
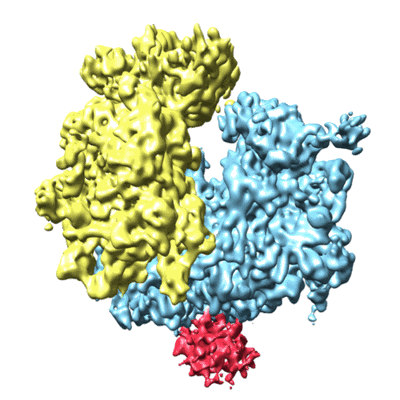
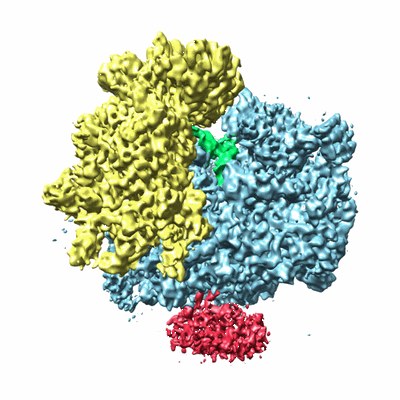
Cryo-EM map of Trigger Factor bound to a translating ribosome
Deeng J, Chan KY, van der Sluis E, Bischoff L, Berninghausen O, Han W, Gumbart J, Schulten K, Beatrix B, Beckmann R
J Mol Biol (2016) 428 pp. 3588-3602 [ Pubmed: 27320387 DOI: doi:10.1016/j.jmb.2016.06.007 ]
Cryo-EM map of Trigger Factor bound to a translating ribosome
Resolution: 7.7 Å
EM Method: Single-particle
Fitted PDBs: 4urd
Q-score: 0.042
Deeng J, Chan KY, van der Sluis E, Bischoff L, Berninghausen O, Han W, Gumbart J, Schulten K, Beatrix B, Beckmann R
J Mol Biol (2016) 428 pp. 3588-3602 [ Pubmed: 27320387 DOI: doi:10.1016/j.jmb.2016.06.007 ]
- Cytosolic 70s ribosome (Complex from Escherichia coli)
- Trigger factor (48 kDa, Protein from Escherichia coli)
- Tnac-stalled-rnc with trigger factor (Sample)
Cryo-EM map of Trigger Factor bound to a translating ribosome
Deeng J, Chan KY, van der Sluis E, Bischoff L, Berninghausen O, Han W, Gumbart J, Schulten K, Beatrix B, Beckmann R
J Mol Biol (2016) 428 pp. 3588-3602 [ Pubmed: 27320387 DOI: doi:10.1016/j.jmb.2016.06.007 ]
Cryo-EM structure of the programmed yeast 80 ribosome bound the Ssh1 complex
Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R
Science (2009) 326 pp. 1369-1373 [ DOI: doi:10.1126/science.1178535 Pubmed: 19933108 ]
- Ssh1 complex (71 kDa, Ligand from Saccharomyces cerevisiae)
- This map represents a yeast 80s ribosome in the unratcheted conformation with the yeast ssh1 complex bound after sorting the entire dataset against an empty (ratcheted) yeast 80s ribosome (4 MDa, Sample)
- Yeast 80s ribosome bound to the yeast ssh1 complex (4 MDa, Complex)
Structure of the ribosome-SecYE complex in the membrane environment
Resolution: 7.1 Å
EM Method: Single-particle
Fitted PDBs: 4v6m
Q-score: 0.145
Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R
Nat Struct Mol Biol (2011) 18 pp. 614-621 [ Pubmed: 21499241 DOI: doi:10.1038/nsmb.2026 ]
Resolution: 8.6 Å
EM Method: Single-particle
Fitted PDBs: 2ww9
Q-score: 0.078
Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R
Science (2009) 326 pp. 1369-1373 [ DOI: doi:10.1126/science.1178535 Pubmed: 19933108 ]
Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R
Science (2009) 326 pp. 1369-1373 [ DOI: doi:10.1126/science.1178535 Pubmed: 19933108 ]
Cryo-EM structures of the idle yeast Ssh1 complex bound to the yeast 80S ribosome
Resolution: 8.8 Å
EM Method: Single-particle
Fitted PDBs: 4v6i, 3izd
Q-score: 0.0565
Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R
Science (2009) 326 pp. 1369-1373 [ DOI: doi:10.1126/science.1178535 Pubmed: 19933108 ]
Resolution: 6.48 Å
EM Method: Single-particle
Fitted PDBs: 2wwb
Q-score: 0.092
Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R
Science (2009) 326 pp. 1369-1373 [ DOI: doi:10.1126/science.1178535 Pubmed: 19933108 ]
- Wheat germ (triticum aestivum) 80 ribosome (4 MDa, Complex)
- Actively translating wheat germ (o. sativa) 80s ribosome programmed with a nascent polypeptide chain containing a p-site trna and the type i signal anchor sequence of dpap-b (dipeptidylaminopeptidase b) bound to the mammalian (canis familiaris) sec61 complex. (4 MDa, Sample)
- Mammalian sec61 complex (70 kDa, Protein from Canis lupus familiaris)