Cryo-EM structure of MraY in complex with analogue 2
Resolution: 2.88 Å
EM Method: Single-particle
Fitted PDBs: 9b70
Q-score: 0.561
Yamamoto K, Sato T, Hao A, Asao K, Kaguchi R, Kusaka S, Ruddarraju RR, Kazamori D, Seo K, Takahashi S, Horiuchi M, Yokota SI, Lee SY, Ichikawa S
Nat Commun (2024) 15 pp. 5085-5085 [ Pubmed: 38877016 DOI: doi:10.1038/s41467-024-49484-7 ]
- Complex of mray aa dimer with nanobody bound (200 kDa, Complex from Aquifex aeolicus VF5)
- Water (18 Da, Ligand)
- Phospho-n-acetylmuramoyl-pentapeptide-transferase (40 kDa, Protein from Aquifex aeolicus VF5)
- Mrayaa nanobody (15 kDa, Protein from Lama glama)
- (2~{s},3~{s})-3-[(2~{s},3~{r},4~{s},5~{r})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-2-[[3-[[[(2~{s})-6-azanyl-2-(hexadecanoylamino)hexanoyl]amino]methyl]phenyl]methylamino]-3-[(2~{s},3~{s},4~{r},5~{r})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propanoic acid (934 Da, Ligand)
Cryo-EM structure of MraY in complex with analogue 3
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 9b71
Q-score: 0.585
Yamamoto K, Sato T, Hao A, Asao K, Kaguchi R, Kusaka S, Ruddarraju RR, Kazamori D, Seo K, Takahashi S, Horiuchi M, Yokota SI, Lee SY, Ichikawa S
Nat Commun (2024) 15 pp. 5085-5085 [ Pubmed: 38877016 DOI: doi:10.1038/s41467-024-49484-7 ]
- Complex of mray aa dimer with nanobody bound (200 kDa, Complex from Aquifex aeolicus VF5)
- Water (18 Da, Ligand)
- Phospho-n-acetylmuramoyl-pentapeptide-transferase (40 kDa, Protein from Aquifex aeolicus VF5)
- Mrayaa nanobody (15 kDa, Protein from Lama glama)
- (2~{s},3~{s})-3-[(2~{s},3~{r},4~{s},5~{r})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-3-[(2~{s},3~{s},4~{r},5~{r})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]-2-[[4-[[[(2~{s})-5-carbamimidamido-2-(hexadecanoylamino)pentanoyl]amino]methyl]phenyl]methylamino]propanoic acid (962 Da, Ligand)
Omicron spike variant XBB with n3130v-Fc
Resolution: 3.44 Å
EM Method: Single-particle
Fitted PDBs: 8i4f
Q-score: 0.302
Hao A, Song W, Li C, Zhang X, Tu C, Wang X, Wang P, Wu Y, Ying T, Sun L
Signal Transduct Target Ther (2023) 8 pp. 269-269 [ DOI: doi:10.1038/s41392-023-01534-0 Pubmed: 37422451 ]
- Spike glycoprotein (143 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Omicron spike with 6m6 (Complex from Severe acute respiratory syndrome coronavirus 2)
- N3130v-fc (42 kDa, Protein from Homo sapiens)
- Xbb (Complex from Homo sapiens)
- Bn03 (Complex)
Omicron spike variant BQ.1.1 with n3130v-Fc
Resolution: 3.68 Å
EM Method: Single-particle
Fitted PDBs: 8i4g
Q-score: 0.266
Hao A, Song W, Li C, Zhang X, Tu C, Wang X, Wang P, Wu Y, Ying T, Sun L
Signal Transduct Target Ther (2023) 8 pp. 269-269 [ DOI: doi:10.1038/s41392-023-01534-0 Pubmed: 37422451 ]
- Spike glycoprotein (143 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- N3130v-fc (Complex)
- Omicron spike with 6m6 (Complex from Severe acute respiratory syndrome coronavirus 2)
- N3130v-fc (42 kDa, Protein from Homo sapiens)
- Bq.1.1 (Complex from Homo sapiens)
Cryo-EM structure of Vibrio cholerae FtsE/FtsX complex
Resolution: 3.51 Å
EM Method: Single-particle
Fitted PDBs: 8tzj
Q-score: 0.471
Structure (2024) 32 pp. 188-199.e5 [ DOI: doi:10.1016/j.str.2023.11.007 Pubmed: 38070498 ]
- Cell division protein ftsx (36 kDa, Protein from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Ftsex (200 kDa, Complex from Vibrio cholerae)
- Cell division atp-binding protein ftse (25 kDa, Protein from Vibrio cholerae)
Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, shortened
Resolution: 3.55 Å
EM Method: Single-particle
Fitted PDBs: 8tzk
Q-score: 0.461
Structure (2024) 32 pp. 188-199.e5 [ DOI: doi:10.1016/j.str.2023.11.007 Pubmed: 38070498 ]
- Cell division protein ftsx (36 kDa, Protein from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Ftsex (200 kDa, Complex from Vibrio cholerae)
- Peptidase m23 (43 kDa, Protein from Vibrio cholerae)
- Cell division atp-binding protein ftse (25 kDa, Protein from Vibrio cholerae)
Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, full-length
Resolution: 3.55 Å
EM Method: Single-particle
Fitted PDBs: 8tzl
Q-score: 0.408
Structure (2024) 32 pp. 188-199.e5 [ DOI: doi:10.1016/j.str.2023.11.007 Pubmed: 38070498 ]
- Cell division protein ftsx (36 kDa, Protein from Vibrio cholerae)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Ftsex (200 kDa, Complex from Vibrio cholerae)
- Peptidase m23 (43 kDa, Protein from Vibrio cholerae)
- Cell division atp-binding protein ftse (25 kDa, Protein from Vibrio cholerae)
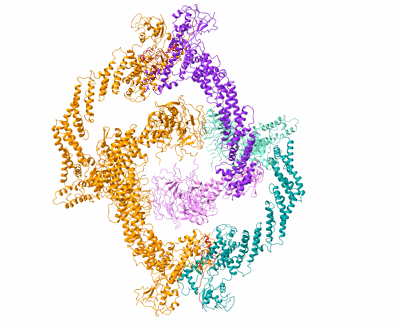
Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 tetrameric complex
Resolution: 7.86 Å
EM Method: Single-particle
Fitted PDBs: 8h33
Q-score: 0.07
Hu Y, Zhang Z, Mao Q, Zhang X, Hao A, Xun Y, Wang Y, Han L, Zhan W, Liu Q, Yin Y, Peng C, Moresco EMY, Chen Z, Beutler B, Sun L
Nat Struct Mol Biol (2024) 31 pp. 336-350 [ DOI: doi:10.1038/s41594-023-01182-6 Pubmed: 38332366 ]
- E3 ubiquitin-protein ligase rbx1 (13 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Cullin-3 (90 kDa, Protein from Homo sapiens)
- Kelch repeat and btb domain-containing protein 2 (71 kDa, Protein from Homo sapiens)
- Kbtbd2-cul3-rbx1 tetrameric complex (Complex from Homo sapiens)
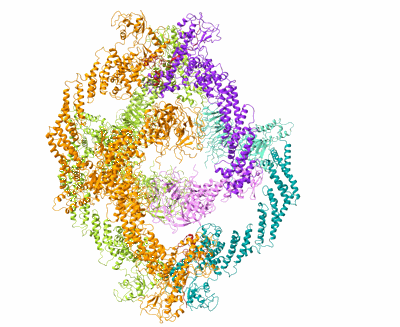
Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex
Resolution: 7.99 Å
EM Method: Single-particle
Fitted PDBs: 8h34
Q-score: 0.088
Hu Y, Zhang Z, Mao Q, Zhang X, Hao A, Xun Y, Wang Y, Han L, Zhan W, Liu Q, Yin Y, Peng C, Moresco EMY, Chen Z, Beutler B, Sun L
Nat Struct Mol Biol (2024) 31 pp. 336-350 [ DOI: doi:10.1038/s41594-023-01182-6 Pubmed: 38332366 ]
- E3 ubiquitin-protein ligase rbx1 (13 kDa, Protein from Homo sapiens)
- Kbtbd2-cul3-rbx1 hexameric complex (Complex from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Cullin-3 (89 kDa, Protein from Homo sapiens)
- Kelch repeat and btb domain-containing protein 2 (71 kDa, Protein from Homo sapiens)
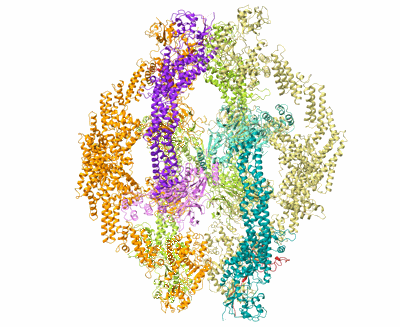
Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 octameric complex
Resolution: 7.41 Å
EM Method: Single-particle
Fitted PDBs: 8h35
Q-score: 0.091
Hu Y, Zhang Z, Mao Q, Zhang X, Hao A, Xun Y, Wang Y, Han L, Zhan W, Liu Q, Yin Y, Peng C, Moresco EMY, Chen Z, Beutler B, Sun L
Nat Struct Mol Biol (2024) 31 pp. 336-350 [ DOI: doi:10.1038/s41594-023-01182-6 Pubmed: 38332366 ]
- E3 ubiquitin-protein ligase rbx1 (12 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Kbtbd2-cul3-rbx1 octameric complex (Complex from Homo sapiens)
- Cullin-3 (89 kDa, Protein from Homo sapiens)
- Kelch repeat and btb domain-containing protein 2 (71 kDa, Protein from Homo sapiens)