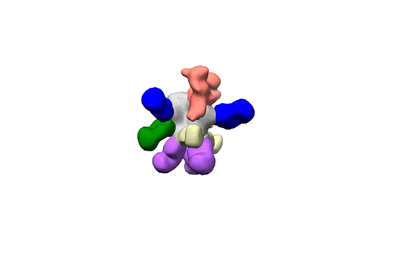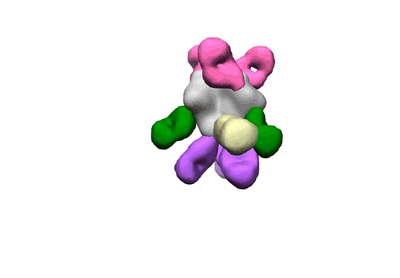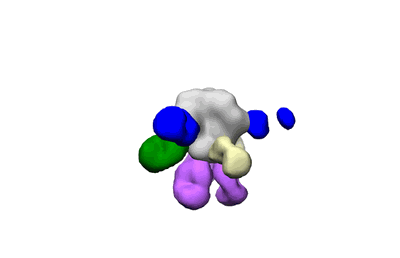Polyclonal Immune complex of Fab binding to BG505 SOSIP.664 from serum of rabbit 5724 at wk 26
Dingens AS, Pratap P, Malone K, Hilton SK, Ketas T, Cottrell CA, Overbaugh J, Moore JP, Klasse PJ, Ward AB, Bloom JD
eLife (2021) 10 [ DOI: doi:10.7554/eLife.64281 Pubmed: 33438580 ]
Polyclonal Immune complex of Fab binding to BG505 SOSIP.664 from serum of rabbit 5727 at wk 26
Dingens AS, Pratap P, Malone K, Hilton SK, Ketas T, Cottrell CA, Overbaugh J, Moore JP, Klasse PJ, Ward AB, Bloom JD
eLife (2021) 10 [ DOI: doi:10.7554/eLife.64281 Pubmed: 33438580 ]
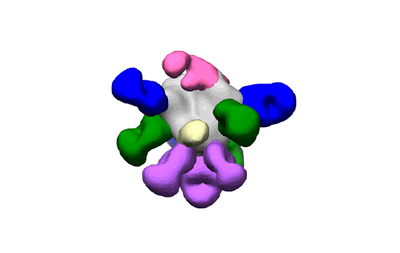
Polyclonal Immune complex of Fab binding to BG505 SOSIP.v4.1 from serum of rabbit 2124 at wk 22
Dingens AS, Pratap P, Malone K, Hilton SK, Ketas T, Cottrell CA, Overbaugh J, Moore JP, Klasse PJ, Ward AB, Bloom JD
eLife (2021) 10 [ DOI: doi:10.7554/eLife.64281 Pubmed: 33438580 ]
Polyclonal Immune complex of Fab binding to BG505 SOSIP.v4.1 from serum of rabbit 2214 at wk 43
Dingens AS, Pratap P, Malone K, Hilton SK, Ketas T, Cottrell CA, Overbaugh J, Moore JP, Klasse PJ, Ward AB, Bloom JD
eLife (2021) 10 [ DOI: doi:10.7554/eLife.64281 Pubmed: 33438580 ]
Polyclonal Immune complex of Fab binding to BG505 SOSIP.v4.1 from serum of rabbit 2423 at wk 18
Dingens AS, Pratap P, Malone K, Hilton SK, Ketas T, Cottrell CA, Overbaugh J, Moore JP, Klasse PJ, Ward AB, Bloom JD
eLife (2021) 10 [ DOI: doi:10.7554/eLife.64281 Pubmed: 33438580 ]
Polyclonal Immune complex of Fab binding to BG505 SOSIP.v4.1 from serum of rabbit 2425 at wk 18
Dingens AS, Pratap P, Malone K, Hilton SK, Ketas T, Cottrell CA, Overbaugh J, Moore JP, Klasse PJ, Ward AB, Bloom JD
eLife (2021) 10 [ DOI: doi:10.7554/eLife.64281 Pubmed: 33438580 ]
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2082
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe Jr JE, Bloom JD
Cell Host Microbe (2021) 29 pp. 44-57.e9 [ DOI: doi:10.1016/j.chom.2020.11.007 Pubmed: 33259788 ]
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2479
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe Jr JE, Bloom JD
Cell Host Microbe (2021) 29 pp. 44-57.e9 [ DOI: doi:10.1016/j.chom.2020.11.007 Pubmed: 33259788 ]
Negative stain EM map of SARS-COV-2 spike protein open RBD (trimer) with Fab COV2-2096
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe Jr JE, Bloom JD
Cell Host Microbe (2021) 29 pp. 44-57.e9 [ DOI: doi:10.1016/j.chom.2020.11.007 Pubmed: 33259788 ]
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2832
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe Jr JE, Bloom JD
Cell Host Microbe (2021) 29 pp. 44-57.e9 [ DOI: doi:10.1016/j.chom.2020.11.007 Pubmed: 33259788 ]