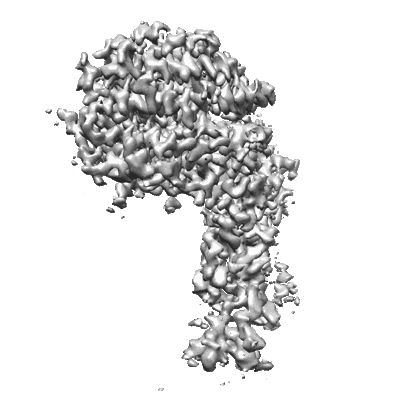
RBD of SARS-CoV2 spike protein with ACE2 decoy
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8jje
Q-score: 0.488
Urano E, Itoh Y, Suzuki T, Sasaki T, Kishikawa JI, Akamatsu K, Higuchi Y, Sakai Y, Okamura T, Mitoma S, Sugihara F, Takada A, Kimura M, Nakao S, Hirose M, Sasaki T, Koketsu R, Tsuji S, Yanagida S, Shioda T, Hara E, Matoba S, Matsuura Y, Kanda Y, Arase H, Okada M, Takagi J, Kato T, Hoshino A, Yasutomi Y, Saito A, Okamoto T
Sci Transl Med (2023) 15 pp. eadi2623-eadi2623 [ DOI: doi:10.1126/scitranslmed.adi2623 Pubmed: 37647387 ]
- Sars-cov2 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Ace2 (Complex from Homo sapiens)
- Sulfate ion (96 Da, Ligand)
- Water (18 Da, Ligand)
- Angiotensin-converting enzyme 2 (73 kDa, Protein from Homo sapiens)
- Spike glycoprotein (138 kDa, Protein from synthetic construct)
- Zinc ion (65 Da, Ligand)
- Rbd of sars-cov2 spike protein with ace2 decoy. (460 kDa, Complex)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
SARS-CoV-2 BA.1 spike with UT28-RD
Resolution: 3.22 Å
EM Method: Single-particle
Fitted PDBs: 8k5h
Q-score: 0.497
Ozawa T, Ikeda Y, Chen L, Suzuki R, Hoshino A, Noguchi A, Kita S, Anraku Y, Igarashi E, Saga Y, Inasaki N, Taminishi S, Sasaki J, Kirita Y, Fukuhara H, Maenaka K, Hashiguchi T, Fukuhara T, Hirabayashi K, Tani H, Kishi H, Niimi H
Structure (2024) 32 pp. 263-272.e7 [ DOI: doi:10.1016/j.str.2023.12.013 Pubmed: 38228146 ]
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ut28k-rd fab light chain (25 kDa, Protein from Homo sapiens)
- Ut28k-rd fab (Complex from Homo sapiens)
- Sars-cov-2 ba.1 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 ba.1 spike protein with ut28k-rd complex (50 kDa, Complex)
- Ut28k-rd fab heavy chain (27 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
SARS-CoV-2 spike in complex with neutralizing antibody NIV-11
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8hgl
Q-score: 0.368
Moriyama S, Anraku Y, Taminishi S, Adachi Y, Kuroda D, Kita S, Higuchi Y, Kirita Y, Kotaki R, Tonouchi K, Yumoto K, Suzuki T, Someya T, Fukuhara H, Kuroda Y, Yamamoto T, Onodera T, Fukushi S, Maeda K, Nakamura-Uchiyama F, Hashiguchi T, Hoshino A, Maenaka K, Takahashi Y
Nat Commun (2023) 14 pp. 4198-4198 [ DOI: doi:10.1038/s41467-023-39890-8 Pubmed: 37452031 ]
- Niv-11 fab heavy chain (23 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Niv-11 fab light chain (23 kDa, Protein from Homo sapiens)
- Niv-11 fab (50 kDa, Complex from Homo sapiens)
- Sars-cov-2 spike glycoprotein (420 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike glycoprotein in complex with niv-11 (570 kDa, Complex)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 focused on RBD and NIV-11 interface
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8hgm
Q-score: 0.459
Moriyama S, Anraku Y, Taminishi S, Adachi Y, Kuroda D, Kita S, Higuchi Y, Kirita Y, Kotaki R, Tonouchi K, Yumoto K, Suzuki T, Someya T, Fukuhara H, Kuroda Y, Yamamoto T, Onodera T, Fukushi S, Maeda K, Nakamura-Uchiyama F, Hashiguchi T, Hoshino A, Maenaka K, Takahashi Y
Nat Commun (2023) 14 pp. 4198-4198 [ DOI: doi:10.1038/s41467-023-39890-8 Pubmed: 37452031 ]
- Niv-11 fab heavy chain (23 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Niv-11 fab light chain (23 kDa, Protein from Homo sapiens)
- Niv-11 fab (50 kDa, Complex from Homo sapiens)
- Sars-cov-2 spike glycoprotein (420 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike glycoprotein in complex with niv-11 (570 kDa, Complex)
Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex
Resolution: 3.08 Å
EM Method: Single-particle
Fitted PDBs: 8j12
Q-score: 0.479
Hino T, Omura SN, Nakagawa R, Togashi T, Takeda SN, Hiramoto T, Tasaka S, Hirano H, Tokuyama T, Uosaki H, Ishiguro S, Kagieva M, Yamano H, Ozaki Y, Motooka D, Mori H, Kirita Y, Kise Y, Itoh Y, Matoba S, Aburatani H, Yachie N, Karvelis T, Siksnys V, Ohmori T, Hoshino A, Nureki O
Cell (2023) 186 pp. 4920-4935.e23 [ Pubmed: 37776859 DOI: doi:10.1016/j.cell.2023.08.031 ]
- Rna (247-mer) (72 kDa, RNA from Acidibacillus sulfooxidans)
- Dna (38-mer) (11 kDa, DNA from Acidibacillus sulfooxidans)
- Magnesium ion (24 Da, Ligand)
- Ascas12f-sgrna-target dna (Complex from Mycolicibacterium mucogenicum)
- Dna (38-mer) (11 kDa, DNA from Acidibacillus sulfooxidans)
- Transposase is605 orfb c-terminal domain-containing protein (49 kDa, Protein from Acidibacillus sulfuroxidans)
- Zinc ion (65 Da, Ligand)
Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA
Resolution: 2.91 Å
EM Method: Single-particle
Fitted PDBs: 8j1j
Q-score: 0.511
Hino T, Omura SN, Nakagawa R, Togashi T, Takeda SN, Hiramoto T, Tasaka S, Hirano H, Tokuyama T, Uosaki H, Ishiguro S, Kagieva M, Yamano H, Ozaki Y, Motooka D, Mori H, Kirita Y, Kise Y, Itoh Y, Matoba S, Aburatani H, Yachie N, Karvelis T, Siksnys V, Ohmori T, Hoshino A, Nureki O
Cell (2023) 186 pp. 4920-4935.e23 [ Pubmed: 37776859 DOI: doi:10.1016/j.cell.2023.08.031 ]
- Dna (38-mer) (11 kDa, DNA from Sulfoacidibacillus thermotolerans)
- Magnesium ion (24 Da, Ligand)
- Transposase is605 orfb c-terminal domain-containing protein (49 kDa, Protein from Sulfoacidibacillus thermotolerans)
- Ascas12f-yham-sgrnas3-5v7-target dna (Complex from Sulfoacidibacillus thermotolerans)
- Dna (38-mer) (11 kDa, DNA from Sulfoacidibacillus thermotolerans)
- Zinc ion (65 Da, Ligand)
- Rna (118-mer) (38 kDa, RNA from Sulfoacidibacillus thermotolerans)
Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA
Resolution: 2.95 Å
EM Method: Single-particle
Fitted PDBs: 8j3r
Q-score: 0.174
Hino T, Omura SN, Nakagawa R, Togashi T, Takeda SN, Hiramoto T, Tasaka S, Hirano H, Tokuyama T, Uosaki H, Ishiguro S, Kagieva M, Yamano H, Ozaki Y, Motooka D, Mori H, Kirita Y, Kise Y, Itoh Y, Matoba S, Aburatani H, Yachie N, Karvelis T, Siksnys V, Ohmori T, Hoshino A, Nureki O
Cell (2023) 186 pp. 4920-4935.e23 [ Pubmed: 37776859 DOI: doi:10.1016/j.cell.2023.08.031 ]
- Magnesium ion (24 Da, Ligand)
- Rna (118-mer) (38 kDa, RNA from Sulfoacidibacillus thermotolerans)
- Ascas12f-hkra (Complex from Sulfoacidibacillus thermotolerans)
- Dna (38-mer) (11 kDa, DNA from Sulfoacidibacillus thermotolerans)
- Zinc ion (65 Da, Ligand)
- Dna (37-mer) (11 kDa, DNA from Sulfoacidibacillus thermotolerans)
- Rna (Complex)
- Transposase is605 orfb c-terminal domain-containing protein (49 kDa, Protein from Sulfoacidibacillus thermotolerans)
- Ascas12f-hkra-sgrnas3-5v7-target dna (Complex from Sulfoacidibacillus thermotolerans)
- Dna (Complex from Sulfoacidibacillus thermotolerans)
SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2)
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7yh7
Q-score: 0.394
Moriyama S, Anraku Y, Taminishi S, Adachi Y, Kuroda D, Kita S, Higuchi Y, Kirita Y, Kotaki R, Tonouchi K, Yumoto K, Suzuki T, Someya T, Fukuhara H, Kuroda Y, Yamamoto T, Onodera T, Fukushi S, Maeda K, Nakamura-Uchiyama F, Hashiguchi T, Hoshino A, Maenaka K, Takahashi Y
Nat Commun (2023) 14 pp. 4198-4198 [ DOI: doi:10.1038/s41467-023-39890-8 Pubmed: 37452031 ]
- Niv-8 fab (50 kDa, Complex from Homo sapiens)
- Sars-cov-2 spike glycoprotein in complex with niv-8 (570 kDa, Complex)
- Sars-cov-2 spike glycoprotein (420 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (132 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Niv-8 fab light chain (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Niv-8 fab heavy chain (13 kDa, Protein from Homo sapiens)