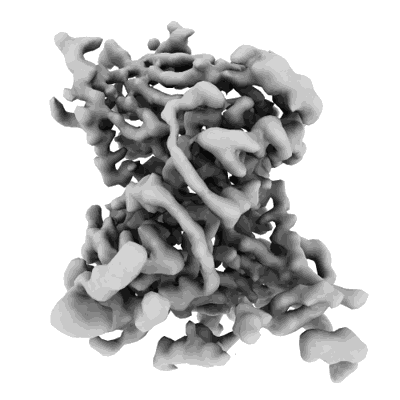
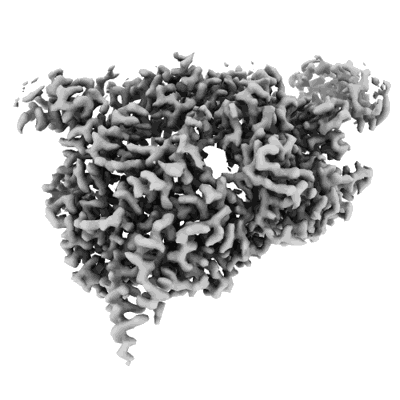
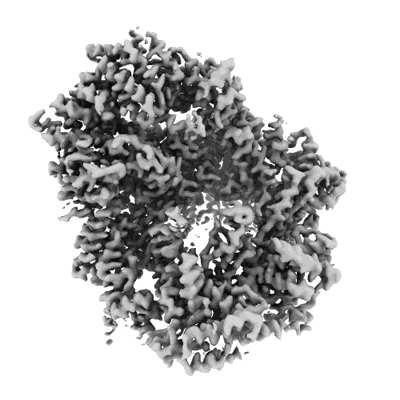
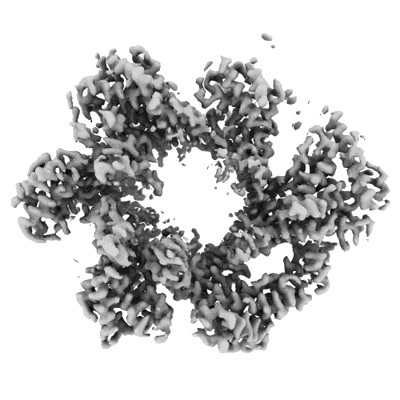Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8ifg
Q-score: 0.441
Wang X, Wang Y, Liu S, Zhang Y, Xu K, Ji L, Kornberg RD, Zhang H
PNAS (2023) 120 pp. e2307598120- [ DOI: doi:10.1073/pnas.2307598120 Pubmed: 37459529 ]
- Chromatin modification-related protein eaf3 (39 kDa, Protein from Schizosaccharomyces pombe (strain 972 / ATCC 24843))
- Water (18 Da, Ligand)
- Cph2 (68 kDa, Protein from Schizosaccharomyces pombe (strain 972 / ATCC 24843))
- Cryo-em structure of the clr6-hdac (clr6s) complex from s. pombe (600 kDa, Complex from Schizosaccharomyces pombe)
- Zinc ion (65 Da, Ligand)
- Rbap48-related wd40 repeat-containing protein prw1 (48 kDa, Protein from Schizosaccharomyces pombe (strain 972 / ATCC 24843))
- Paired amphipathic helix protein pst2 (125 kDa, Protein from Schizosaccharomyces pombe (strain 972 / ATCC 24843))
- Histone deacetylase clr6 (45 kDa, Protein from Schizosaccharomyces pombe (strain 972 / ATCC 24843))
- Cph1 (45 kDa, Protein from Schizosaccharomyces pombe (strain 972 / ATCC 24843))
Clr6 HDAC (Clr6S)-nucleosome complex
Wang X, Wang Y, Liu S, Zhang Y, Xu K, Ji L, Kornberg RD, Zhang H
PNAS (2023) 120 pp. e2307598120-e2307598120 [ DOI: doi:10.1073/pnas.2307598120 Pubmed: 37459529 ]
Rpd3S-nucleosome (H3K36me3 modified)
Wang X, Wang Y, Liu S, Zhang Y, Xu K, Ji L, Kornberg RD, Zhang H
PNAS (2023) 120 pp. e2307598120-e2307598120 [ DOI: doi:10.1073/pnas.2307598120 Pubmed: 37459529 ]
Krishna Kumar K, Robertson MJ, Thadhani E, Wang H, Suomivuori CM, Powers AS, Ji L, Nikas SP, Dror RO, Inoue A, Makriyannis A, Skiniotis G, Kobilka B
Nat Commun (2023) 14 [ DOI: 10.1038/s41467-023-37864-4 Pubmed: 37160876 ]
Image sets:
Xia Y, Sonneville R, Jenkyn-Bedford M, Ji L, Alabert C, Hong Y, Yeeles JTP, Labib KPM
Science (2023) 381 pp. eadi4932-eadi4932 [ DOI: doi:10.1126/science.adi4932 Pubmed: 37590372 ]
Xia Y, Sonneville R, Jenkyn-Bedford M, Ji L, Alabert C, Hong Y, Yeeles JTP, Labib KPM
Science (2023) 381 pp. eadi4932-eadi4932 [ DOI: doi:10.1126/science.adi4932 Pubmed: 37590372 ]
Xia Y, Sonneville R, Jenkyn-Bedford M, Ji L, Alabert C, Hong Y, Yeeles JTP, Labib KPM
Science (2023) 381 pp. eadi4932-eadi4932 [ DOI: doi:10.1126/science.adi4932 Pubmed: 37590372 ]
Xia Y, Sonneville R, Jenkyn-Bedford M, Ji L, Alabert C, Hong Y, Yeeles JTP, Labib KPM
Science (2023) 381 pp. eadi4932-eadi4932 [ DOI: doi:10.1126/science.adi4932 Pubmed: 37590372 ]
Xia Y, Sonneville R, Jenkyn-Bedford M, Ji L, Alabert C, Hong Y, Yeeles JTP, Labib KPM
Science (2023) 381 pp. eadi4932-eadi4932 [ DOI: doi:10.1126/science.adi4932 Pubmed: 37590372 ]
Xia Y, Sonneville R, Jenkyn-Bedford M, Ji L, Alabert C, Hong Y, Yeeles JTP, Labib KPM
Science (2023) 381 pp. eadi4932-eadi4932 [ DOI: doi:10.1126/science.adi4932 Pubmed: 37590372 ]