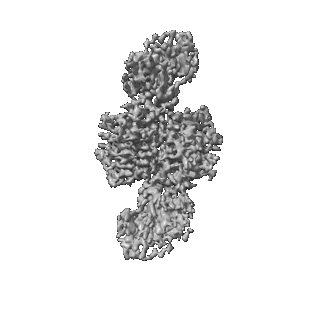
Caspase-1 complex with interleukin-18
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8sv1
Q-score: 0.353
Dong Y, Bonin JP, Devant P, Liang Z, Sever AIM, Mintseris J, Aramini JM, Du G, Gygi SP, Kagan JC, Kay LE, Wu H
Immunity (2024) 57 pp. 1533-1548.e10 [ DOI: doi:10.1016/j.immuni.2024.04.015 Pubmed: 38733997 ]
- Interleukin-18 (21 kDa, Protein from Homo sapiens)
- Caspase-1 (16 kDa, Protein from Homo sapiens)
- Protein complex with interleukin (100 MDa, Complex from Homo sapiens)
- Caspase-1 (10 kDa, Protein from Homo sapiens)
Structure of Ran bound to RCC1 and the nucleosome core particle (composite map)
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8ux1
Q-score: 0.609
Huang SK, Rubinstein JL, Kay LE
To Be Published
- 153-bp widom 601 dna forward strand (47 kDa, DNA from synthetic construct)
- Regulator of chromosome condensation (44 kDa, Protein from Homo sapiens)
- Histone h2b (13 kDa, Protein from Drosophila melanogaster)
- Histone h3 (15 kDa, Protein from Drosophila melanogaster)
- 153-bp widom 601 dna reverse strand (46 kDa, DNA from synthetic construct)
- Histone h2a (13 kDa, Protein from Drosophila melanogaster)
- Histone h4 (11 kDa, Protein from Drosophila melanogaster)
- Ran-rcc1-ncp complex (Complex from Homo sapiens)
- Gtp-binding nuclear protein ran (24 kDa, Protein from Homo sapiens)
Cryo-EM maps of the NCP-RCC1-Ran complex with multiple binding states
Focused-refinement map of the RCC1-Ran region from the NCP-RCC1-Ran complex
Cryo-EM map of the Unmodified nucleosome core particle in 100 mM KCl with local resolution values
Nosella ML, Kim TH, Huang SK, Harkness RW, Goncalves M, Pan A, Tereshchenko M, Vahidi S, Rubinstein JL, Lee HO, Forman-Kay JD, Kay LE
Mol Cell (2024) 84 pp. 429-446.e17 [ DOI: doi:10.1016/j.molcel.2023.12.019 Pubmed: 38215753 ]
Cryo-EM map of the PARylated nucleosome core particle in 100 mM KCl with local resolution values
Nosella ML, Kim TH, Huang SK, Harkness RW, Goncalves M, Pan A, Tereshchenko M, Vahidi S, Rubinstein JL, Lee HO, Forman-Kay JD, Kay LE
Mol Cell (2024) 84 pp. 429-446.e17 [ DOI: doi:10.1016/j.molcel.2023.12.019 Pubmed: 38215753 ]
Cryo-EM map of the Unmodified nucleosome core particle in 5 mM KCl with local resolution values
Nosella ML, Kim TH, Huang SK, Harkness RW, Goncalves M, Pan A, Tereshchenko M, Vahidi S, Rubinstein JL, Lee HO, Forman-Kay JD, Kay LE
Mol Cell (2024) 84 pp. 429-446.e17 [ DOI: doi:10.1016/j.molcel.2023.12.019 Pubmed: 38215753 ]
Cryo-EM map of the PARylated nucleosome core particle in 5 mM KCl with local resolution values
Nosella ML, Kim TH, Huang SK, Harkness RW, Goncalves M, Pan A, Tereshchenko M, Vahidi S, Rubinstein JL, Lee HO, Forman-Kay JD, Kay LE
Mol Cell (2024) 84 pp. 429-446.e17 [ DOI: doi:10.1016/j.molcel.2023.12.019 Pubmed: 38215753 ]
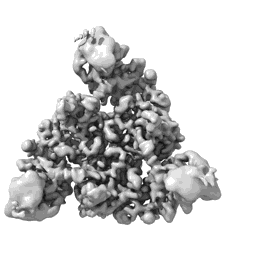
Client-bound structure of a DegP trimer within a 12mer cage
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8f0a
Q-score: 0.513
Harkness RW, Ripstein ZA, Di Trani JM, Kay LE
J Am Chem Soc (2023) 145 pp. 13015-13026 [ DOI: doi:10.1021/jacs.2c11849 Pubmed: 37282495 ]
- Periplasmic serine endoprotease degp (7 kDa, Protein from Escherichia coli (strain K12))
- Telomeric repeat-binding factor 1 (3 kDa, Protein from Homo sapiens)
- Complex of a degp trimer and the client protein htrf1 from a 12mer cage structure (Complex from Escherichia coli (strain K12))
- Periplasmic serine endoprotease degp (36 kDa, Protein from Escherichia coli (strain K12))
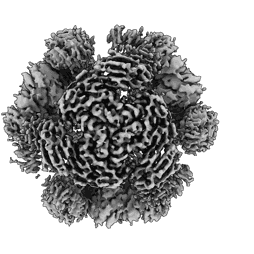
Structure of a 12mer DegP cage bound to the client protein hTRF1
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8f0u
Q-score: 0.359
Harkness RW, Ripstein ZA, Di Trani JM, Kay LE
J Am Chem Soc (2023) 145 pp. 13015-13026 [ DOI: doi:10.1021/jacs.2c11849 Pubmed: 37282495 ]
- Periplasmic serine endoprotease degp (7 kDa, Protein from Escherichia coli (strain K12))
- Telomeric repeat-binding factor 1 (3 kDa, Protein from Homo sapiens)
- Structure of a 12mer degp cage bound to the client protein htrf1 (Complex from Escherichia coli (strain K12))
- Periplasmic serine endoprotease degp (36 kDa, Protein from Escherichia coli (strain K12))