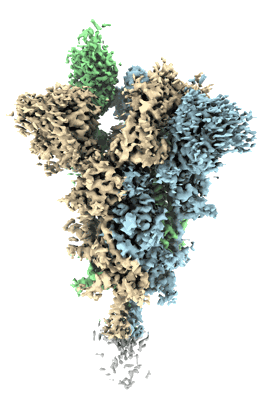
Cryo-EM structure of PGDM1400 Fab bound to HIV-1 BG505 DS-SOSIP.664 Env trimer
Resolution: 3.44 Å
EM Method: Single-particle
Fitted PDBs: 9d1w
Q-score: 0.483
Mason RD, Zhang B, Morano NC, Shen CH, McKee K, Heimann A, Du R, Nazzari AF, Hodges S, Kanai T, Lin BC, Louder MK, Doria-Rose NA, Zhou T, Shapiro L, Roederer M, Kwong PD, Gorman J
Cell Rep (2025) 44 pp. 115223-115223 [ Pubmed: 39826122 DOI: doi:10.1016/j.celrep.2024.115223 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Hiv-1 bg505 ds-sosip glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Pgdm1400 heavy chain (26 kDa, Protein from Homo sapiens)
- Pgdm1400 fab with hiv-1 bg505 ds-sosip.664 env trimer (Complex from Human immunodeficiency virus 1)
- Hiv-1 bg505 ds-sosip gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
- Pgdm1400 light chain (24 kDa, Protein from Homo sapiens)
Cryo-EM structure of PGT145 R100aS Fab bound to HIV-1 BG505 DS-SOSIP.664 Env trimer
Resolution: 3.41 Å
EM Method: Single-particle
Fitted PDBs: 9d3d
Q-score: 0.474
Mason RD, Zhang B, Morano NC, Shen CH, McKee K, Heimann A, Du R, Nazzari AF, Hodges S, Kanai T, Lin BC, Louder MK, Doria-Rose NA, Zhou T, Shapiro L, Roederer M, Kwong PD, Gorman J
Cell Rep (2025) 44 pp. 115223-115223 [ Pubmed: 39826122 DOI: doi:10.1016/j.celrep.2024.115223 ]
- Phosphate ion (94 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Pgt145 r100as heavy chain (26 kDa, Protein from Homo sapiens)
- Bg505 ds-sosip glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Pgt145 r100as fab with hiv-1 bg505 ds-sosip.664 env trimer (Complex from Human immunodeficiency virus 1)
- Hiv-1 bg505 ds-sosip gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
- Pgt145 light chain (23 kDa, Protein from Homo sapiens)
Cryo-EM structure of SIVmac239 SOS-2P Env trimer in complex with human bNAb PGT145
Resolution: 4.12 Å
EM Method: Single-particle
Fitted PDBs: 8dvd
Q-score: 0.322
Gorman J, Wang C, Mason RD, Nazzari AF, Welles HC, Zhou T, Bess Jr JW, Bylund T, Lee M, Tsybovsky Y, Verardi R, Wang S, Yang Y, Zhang B, Rawi R, Keele BF, Lifson JD, Liu J, Roederer M, Kwong PD
Nat Struct Mol Biol (2022) 29 pp. 1080-1091 [ DOI: doi:10.1038/s41594-022-00852-1 Pubmed: 36344847 ]
- Sivmac239 sos-2p in complex with pgt145 (Complex from Simian immunodeficiency virus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Pgt145 heavy (26 kDa, Protein from Homo sapiens)
- Envelope glycoprotein gp160 (57 kDa, Protein from Simian immunodeficiency virus)
- Pgt145 light (23 kDa, Protein from Homo sapiens)
- Envelope glycoprotein gp160 (16 kDa, Protein from Simian immunodeficiency virus)
- Alpha-d-mannopyranose (180 Da, Ligand)
SIV E660.CR54 SOS-2P Env Trimer with ITS92.02
Resolution: 4.32 Å
EM Method: Single-particle
Fitted PDBs: 8dua
Q-score: 0.249
Gorman J, Wang C, Mason RD, Nazzari AF, Welles HC, Zhou T, Bess Jr JW, Bylund T, Lee M, Tsybovsky Y, Verardi R, Wang S, Yang Y, Zhang B, Rawi R, Keele BF, Lifson JD, Liu J, Roederer M, Kwong PD
Nat Struct Mol Biol (2022) 29 pp. 1080-1091 [ DOI: doi:10.1038/s41594-022-00852-1 Pubmed: 36344847 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Envelope glycoprotein gp41 (16 kDa, Protein from Simian immunodeficiency virus)
- Envelope glycoprotein gp120 (59 kDa, Protein from Simian immunodeficiency virus)
- Siv e660.cr54 sos-2p with its92.02 (Complex from Influenza A virus)
- Its92.02 light chain (23 kDa, Protein from Macaca mulatta)
- Its92.02 heavy chain (24 kDa, Protein from Macaca mulatta)
In-situ structure of SIV trimer
Gorman J, Wang C, Mason RD, Nazzari AF, Welles HC, Zhou T, Bess Jr JW, Bylund T, Lee M, Tsybovsky Y, Verardi R, Wang S, Yang Y, Zhang B, Rawi R, Keele BF, Lifson JD, Liu J, Roederer M, Kwong PD
Nat Struct Mol Biol (2022) 29 pp. 1080-1091 [ DOI: doi:10.1038/s41594-022-00852-1 Pubmed: 36344847 ]
in situ SIVmac239-Env trimers on the surface of AT-2-inactivated virions
Gorman J, Wang C, Mason RD, Nazzari AF, Welles HC, Zhou T, Bess Jr JW, Bylund T, Lee M, Tsybovsky Y, Verardi R, Wang S, Yang Y, Zhang B, Rawi R, Keele BF, Lifson JD, Liu J, Roederer M, Kwong PD
Nat Struct Mol Biol (2022) 29 pp. 1080-1091 [ DOI: doi:10.1038/s41594-022-00852-1 Pubmed: 36344847 ]
in situ SIVmac239-Env trimers on the surface of AT-2-inactivated virions
Gorman J, Wang C, Mason RD, Nazzari AF, Welles HC, Zhou T, Bess Jr JW, Bylund T, Lee M, Tsybovsky Y, Verardi R, Wang S, Yang Y, Zhang B, Rawi R, Keele BF, Lifson JD, Liu J, Roederer M, Kwong PD
Nat Struct Mol Biol (2022) 29 pp. 1080-1091 [ DOI: doi:10.1038/s41594-022-00852-1 Pubmed: 36344847 ]
In-situ structure of SIVmac239-Env trimers with ITS90.03 associated
Gorman J, Wang C, Mason RD, Nazzari AF, Welles HC, Zhou T, Bess Jr JW, Bylund T, Lee M, Tsybovsky Y, Verardi R, Wang S, Yang Y, Zhang B, Rawi R, Keele BF, Lifson JD, Liu J, Roederer M, Kwong PD
Nat Struct Mol Biol (2022) 29 pp. 1080-1091 [ DOI: doi:10.1038/s41594-022-00852-1 Pubmed: 36344847 ]
SIV mac239 SOS-2P K169T Env trimer with bNAb PGT145 and jacalin
Gorman J, Wang C, Mason RD, Nazzari AF, Welles HC, Zhou T, Bess Jr JW, Bylund T, Lee M, Tsybovsky Y, Verardi R, Wang S, Yang Y, Zhang B, Rawi R, Keele BF, Lifson JD, Liu J, Roederer M, Kwong PD
Nat Struct Mol Biol (2022) 29 pp. 1080-1091 [ DOI: doi:10.1038/s41594-022-00852-1 Pubmed: 36344847 ]
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Resolution: 3.29 Å
EM Method: Single-particle
Fitted PDBs: 7tb4
Q-score: 0.406
Wang L, Zhou T, Zhang Y, Yang ES, Schramm CA, Shi W, Pegu A, Oloninyi OK, Ransier A, Darko S, Narpala SR, Hatcher C, Martinez DR, Tsybovsky Y, Phung E, Abiona OM, Cale EM, Chang LA, Corbett KS, DiPiazza AT, Gordon IJ, Leung K, Liu T, Mason RD, Nazzari A, Novik L, Olia AS, Doria-Rose NA, Stephens T, Stringham CD, Talana CA, Teng IT, Wagner D, Widge AT, Zhang B, Roederer M, Ledgerwood JE, Ruckwardt TJ, Gaudinski MR, Baric RS, Graham BS, McDermott AB, Douek DC, Kwong PD, Mascola JR, Sullivan NJ, Misasi J
bioRxiv (2021) [ Pubmed: 33655252 DOI: doi:10.1101/2021.02.25.432969 ]