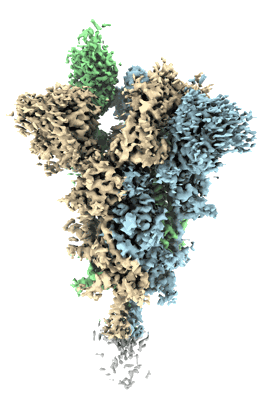
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Resolution: 3.29 Å
EM Method: Single-particle
Fitted PDBs: 7tb4
Q-score: 0.406
Wang L, Zhou T, Zhang Y, Yang ES, Schramm CA, Shi W, Pegu A, Oloninyi OK, Ransier A, Darko S, Narpala SR, Hatcher C, Martinez DR, Tsybovsky Y, Phung E, Abiona OM, Cale EM, Chang LA, Corbett KS, DiPiazza AT, Gordon IJ, Leung K, Liu T, Mason RD, Nazzari A, Novik L, Olia AS, Doria-Rose NA, Stephens T, Stringham CD, Talana CA, Teng IT, Wagner D, Widge AT, Zhang B, Roederer M, Ledgerwood JE, Ruckwardt TJ, Gaudinski MR, Baric RS, Graham BS, McDermott AB, Douek DC, Kwong PD, Mascola JR, Sullivan NJ, Misasi J
bioRxiv (2021) [ Pubmed: 33655252 DOI: doi:10.1101/2021.02.25.432969 ]
Structure of SARS-CoV-2 spike at pH 4.5
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 7jwy
Q-score: 0.612
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike at ph 4.5 (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Water (18 Da, Ligand)
Structure of SARS-CoV-2 spike at pH 4.0
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 6xlu
Q-score: 0.63
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Water (18 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Consensus structure of SARS-CoV-2 spike at pH 5.5
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 6xm0
Q-score: 0.583
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6xm3
Q-score: 0.562
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6xm4
Q-score: 0.559
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 6xm5
Q-score: 0.521
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)